Volume 30, Number 10—October 2024
Dispatch
Oxacillinase-484–Producing Enterobacterales, France, 2018–2023
Abstract
We examined the emergence and characteristics of oxacillinase-484–producing Enterobacterales in France during 2012–2023. Genomic analysis identified 2 predominant sequence types in Escherichia coli: ST410 and ST1722. Plasmid analysis revealed that blaOXA-484 genes were carried mostly on an IncX3-type plasmid associated with genetic elements including insertion sequences IS3000 and ISKpn19.
Carbapenemase-producing Enterobacterales (CPEs) pose a considerable threat to public health because of antimicrobial resistance. In France, the most prevalent carbapenemases are OXA-48–like type. More than 55 variants of OXA-48–like enzymes have been identified (1); OXA-48, OXA-181, and OXA-244 are most prevalent, but OXA-484 has been increasingly identified. OXA-484 differs from OXA-48 by 5 amino acid substitutions (Thr104Ala, Asn110Asp, Glu168Gln, Ser171Ala, Arg214Gly) and from OXA-181 by a single amino acid at position 214 within the β5-β6 loop (2). Most prior reports have described OXA-484 in Escherichia coli (3–6), but some reports also have identified it in K. pneumoniae (2,7), K. aerogenes (6), and K. variicola (8). Reported OXA-484–producing E. coli was noted to belong mainly to the sequence type (ST) 410 (3,5,6), and blaOXA-484 genes were carried mainly 51-kb IncX-type plasmids (3,4,6). Given the similar genetic background of blaOXA-484 and blaOXA-181, several studies suggested that blaOXA-484 could directly derive from blaOXA-181 (3,8). We used whole-genome sequencing to decipher the epidemiology of OXA-484–producing Enterobacterales in France during 2012–2023.
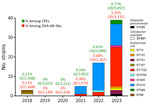
The French National Reference Center (Le Kremlin-Bicêtre, France) receives bacterial strains sent by microbiology laboratories throughout France to analyze for suspected carbapenemase production. The center received 64 clinical, nonduplicate OXA-484–producing isolates during 2012–2023 (Appendix 1 Table 1). We identified OXA-484 producers in 3 isolates from 2018 (0.11% of CPEs, 0.16% of OXA-48–like), 6 from 2021 (0.24% of CPEs, 0.38% of OXA-48–like), 16 from 2022 (0.41% of CPEs, 0.68% of OXA-48–like), and 39 from 2023 (0.77% of CPEs, 1.24% of OXA-48–like) (Figure 1). All OXA-484 producers were E. coli, except 1 K. pneumoniae and 1 Citrobacter youngae. Short-read next-generation sequencing (Appendix 2) showed that the C. youngae isolate belonged to ST491 and the K. pneumoniae isolate to ST268. We identified 10 single STs among the 62 E. coli isolates; most were ST410 (n = 29) and ST1722 (n = 23).
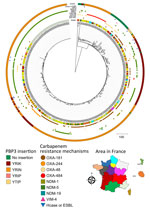
To compare strains belonging to the 2 predominant sequence types (ST410 and ST1722), we constructed a matrix of single-nucleotide polymorphisms (SNPs) in SNIppy version 4.6.0 (ILRI-CGIAR, https://hpc.ilri.cgiar.org). We found 2 ST410 strains (399A8 and 415H3), isolated in the same region, were notably distinct from the others (>1,000 SNPs with the remaining strains) (Appendix 2 Figure 1, panel A). For the other strains, all isolated from different areas, we observed a maximum of 57 SNPs between any 2 strains. We constructed a phylogenetic tree that included all E. coli ST410 strains received at the F-NRC (n = 459). Except for 399A8 and 415H3, the OXA-484 producers formed a distinct cluster among E. coli ST410 (Figure 2).
We observed a maximum of 35 SNPs among the OXA-484–producing E. coli ST1722 (Appendix 2 Figure 1, panel B); all OXA-484 producers clustered inside E. coli ST1722 (n = 43) (Appendix 2 Figure 2). Mapping of short-read data by using CLC Genomics Workbench (QIAGEN, https://www.qiagen.com) onto previously published plasmids (GenBank accession nos. NZ_OP594535, CP058621, CP076530, OP594534, NZ_JANQAU010000004, and CP076530) revealed a high nucleotide identity in most strains. In E. coli, plasmids harboring blaOXA-484 appeared similar to the IncX3 described by Moser et al. (3), with the exception of 2 strains belonging to ST1722, in which plasmids were closed to the IncFIA-like/IncFIB-like/IncFII-type plasmid reported by Findlay (5) (Appendix 1 Table 2). In addition, in C. youngae and E. coli 172D10 strains, the blaOXA-484 gene was located on a new 58,440 pb IncP1 plasmid. For strains having short reads that did not map accurately to described plasmids (query cover <90%, n = 10; Appendix 1 Table 2), we performed long-read sequencing as previously described (9) to reconstruct these plasmids using MinION technology (Oxford Nanopore Technologies, https://nanoporetech.com). The sizes of blaOXA-484–carrying plasmids were 18,731–162,425 bp; 7/10 possessed replicases close to colKP3-like, IncFIA-like, IncFIB-like, and IncFII, and 3 contained only colKP3-like. Some plasmids carried other resistance genes, notably qnrS1 (9/10 plasmids) and mph(A) (4/10 plasmids) (Appendix 1 Table 3).
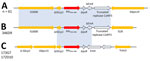
We analyzed the close genetic context of blaOXA-484 and found the same genetic environment for 62/64 strains (Figure 3). For strain 346D9, the close genetic environment was slightly smaller, with the IS3000 upstream and the truncated ColKP3 replicase downstream and an IS26 instead of the ISKpn19. For the blaOXA-484 carried by the IncP1-type plasmid, the genetic environment was totally different, with various truncated IS elements (Figure 3). We found no homologous sequences associated with OXA-48–like or any other carbapenemase-encoding gene.
We determined MICs of various antimicrobial drugs for the entire collection (Table; Appendix 1 Table 4). As expected, OXA-484 producers exhibited low MICs for the 3 carbapenems and were susceptible to imipenem/relebactam and meropenem/vaborbactam combinations (with no inhibitory role of relebactam and vaborbactam). Given that most OXA-484 producers coproduce an extended-spectrum – β-lactamase (CTX-M-14 or CTX-M-15) or an acquired cephalosporinase (mostly CMY-42), those we analyzed were found to be resistant to third- and fourth-generation cephalosporins and to aztreonam, but they remained susceptible to ceftazidime/avibactam and aztreonam/avibactam. All strains were susceptible to colistin (Table).
ST410 E. coli strains possess a 4-aa insertion in penicillin-binding protein 3 (e.g., YRIN motif) (10) that confers decreased susceptibility to certain antibiotics (e.g., ceftazidime and aztreonam). In our collection, all OXA-484–producing ST410 E. coli strains possessed the YRIN insertion in penicillin-binding protein 3 (Figure 2) and exhibited higher MICs for aztreonam/avibactam and ceftazidime/avibactam compared with strains belonging to other sequence types (Appendix 2 Figure 3). In contrast, strains belonging to ST1722 exhibited very low MICs.
A finding of low MICs of temocillin in OXA-484 producers compared with OXA-48 producers aligned with previous reports regarding OXA-244 producers (11) that exhibited the same Arg214Gly mutation within the β5–β6 loop. Because increased susceptibility to temocillin was responsible for screening issues of OXA-244 (11), we evaluated the accuracy of several screening media to detect OXA-484 producers. We selected 46 OXA-484 producers with varying temocillin MICs alongside control strains producing OXA-48 or other carbapenemases. We streaked the strains with 10 µL of 0.5 McFarland bacterial suspension onto 2 commercially available CPE screening media (ChromID CARBA SMART [bioMérieux, https://www.biomerieux.com] and mSuperCARBA [Mast Diagnostic, https://www.mast-group.com/fr]). As expected, after 16–24 hours’ incubation, only 2 strains cultured on the CARB-side and only 11 on the OXA-side of the ChromID CARBA SMART medium. Those 11 isolates displayed temocillin MICs ≥128 mg/L. The mSuperCARBA agar yielded positive results for all OXA-484 producers.
As previously reported for OXA-244 producers (11), we found that the prevalence of OXA-484 producers is likely underestimated because of detection failure related to a lack of sensitivity of ChromID CARBA SMART, the most common screening medium in France. Whereas mSuperCARBA agar appears to be well suited, this medium possesses lower specificity, leading to a rise in additional confirmatory tests (12).
Most previously reported OXA-484 producers belong to E. coli ST410, a widely distributed, high-risk clone (13) that usually exhibits reduced susceptibility to multiple antimicrobial drugs, complicating treatment options. Given the number of SNPs between all OXA-484–producing E. coli ST410 isolates of our collection and their geographic distribution, those strains do not originate from a single outbreak. However, OXA-484–producing E. coli ST410 strains clustered together, suggesting the circulation of a single clone in France during 2018–2023. In addition, most OXA-484–producing E. coli belonged to ST1722. This clone has not been recognized for its prevalence nor for its role in the spread of carbapenem resistance but has been associated with the production of extended-spectrum β-lactamases in France (14).
We found blaOXA-484 genes localized on different plasmids but with a common close genetic environment characterized by IS3000 upstream and ISKpn19 downstream. The ST410 E. coli carried blaOXA-484 on a 51-kb IncX3 plasmid similar to the one described by Moser et al. (4), whereas the ST1722 E. coli carried it on an IncF plasmid similar to the one described by Findlay et al (6). The genetic environment of blaOXA-484 described on IncX3 or IncF-type plasmids is close to that of blaOXA-181, likely the result of a mutation in blaOXA-181 (8). However, the genetic environment observed around the blaOXA-484 gene located on the IncP1-type plasmid does not match any known structures of other blaOXA-48–like genes, questioning its potential origin. In summary, the growing prevalence of OXA-484 producers highlight the urgent need for improved surveillance of these pathogens.
Dr. Emeraud is an assistant professor at the Institut National de la Santé et de la Recherche Médicale. Her primary research interests include epidemiology, genetics, and biochemistry of β-lactamases in gram-negative bacteria.
References
- Naas T, Oueslati S, Bonnin RA, Dabos ML, Zavala A, Dortet L, et al. Beta-lactamase database (BLDB) - structure and function. J Enzyme Inhib Med Chem. 2017;32:917–9. DOIPubMedGoogle Scholar
- Findlay J, Hopkins KL, Loy R, Doumith M, Meunier D, Hill R, et al. OXA-48-like carbapenemases in the UK: an analysis of isolates and cases from 2007 to 2014. J Antimicrob Chemother. 2017;72:1340–9. DOIPubMedGoogle Scholar
- Sommer J, Gerbracht KM, Krause FF, Wild F, Tietgen M, Riedel-Christ S, et al. OXA-484, an OXA-48-type carbapenem-hydrolyzing class D β-lactamase from Escherichia coli. Front Microbiol. 2021;12:
660094 . DOIPubMedGoogle Scholar - Moser AI, Campos-Madueno EI, Sendi P, Perreten V, Keller PM, Ramette A, et al. Repatriation of a patient with COVID-19 contributed to the importation of an emerging carbapenemase producer. J Glob Antimicrob Resist. 2021;27:267–72. DOIPubMedGoogle Scholar
- Hooban B, Fitzhenry K, O’Connor L, Miliotis G, Joyce A, Chueiri A, et al. A longitudinal survey of antibiotic-resistant Enterobacterales in the Irish environment, 2019-2020. Sci Total Environ. 2022;828:
154488 . DOIPubMedGoogle Scholar - Findlay J, Duran JB, Poirel L, Nordmann P. Emergence of OXA-484, an OXA-48-type beta-lactamase, in Switzerland. J Glob Antimicrob Resist. 2023;32:131–3. DOIPubMedGoogle Scholar
- Ramsamy Y, Mlisana KP, Amoako DG, Abia ALK, Ismail A, Allam M, et al. Mobile genetic elements-mediated Enterobacterales-associated carbapenemase antibiotic resistance genes propagation between the environment and humans: A One Health South African study. Sci Total Environ. 2022;806:
150641 . DOIPubMedGoogle Scholar - Ge H, Qiao J, Xu H, Liu R, Zhao J, Chen R, et al. Emergence of OXA-484-Producing Klebsiella variicola in China. Infect Drug Resist. 2023;16:1767–75. DOIPubMedGoogle Scholar
- Girlich D, Bonnin RA, Proust A, Naas T, Dortet L. Undetectable production of the VIM-1 carbapenemase in an Atlantibacter hermannii clinical isolate. Front Microbiol. 2021;12:
741972 . DOIPubMedGoogle Scholar - Sato T, Ito A, Ishioka Y, Matsumoto S, Rokushima M, Kazmierczak KM, et al. Escherichia coli strains possessing a four amino acid YRIN insertion in PBP3 identified as part of the SIDERO-WT-2014 surveillance study. JAC Antimicrob Resist. 2020;2:
dlaa081 . DOIPubMedGoogle Scholar - Emeraud C, Biez L, Girlich D, Jousset AB, Naas T, Bonnin RA, et al. Screening of OXA-244 producers, a difficult-to-detect and emerging OXA-48 variant? J Antimicrob Chemother. 2020;75:2120–3. DOIPubMedGoogle Scholar
- Girlich D, Anglade C, Zambardi G, Nordmann P. Comparative evaluation of a novel chromogenic medium (chromID OXA-48) for detection of OXA-48 producing Enterobacteriaceae. Diagn Microbiol Infect Dis. 2013;77:296–300. DOIPubMedGoogle Scholar
- Ba X, Guo Y, Moran RA, Doughty EL, Liu B, Yao L, et al. Global emergence of a hypervirulent carbapenem-resistant Escherichia coli ST410 clone. Nat Commun. 2024;15:494. DOIPubMedGoogle Scholar
- Muller A, Gbaguidi-Haore H, Cholley P, Hocquet D, Sauget M, Bertrand X. Hospital-diagnosed infections with Escherichia coli clonal group ST131 are mostly acquired in the community. Sci Rep. 2021;11:5702. DOIPubMedGoogle Scholar
Figures
Table
Cite This ArticleOriginal Publication Date: September 18, 2024
Table of Contents – Volume 30, Number 10—October 2024
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Laurent Dortet, Service de Bactériologie-Hygiène, Hôpital de Bicêtre, 78 rue du Général Leclerc, 94275 Le Kremlin-Bicêtre Cedex, France
Top