Volume 8, Number 4—April 2002
Dispatch
Salmonella enterica Serotype Typhimurium DT 104 Antibiotic Resistance Genomic Island I in Serotype Paratyphi B
Abstract
We have identified Salmonella genomic island I (SGI1) in an isolate of Salmonella enterica serotype Paratyphi B. This antibiotic-resistance gene cluster, which confers multidrug resistance, has been previously identified in S. enterica serotype Typhimurium phage types DT 104 and DT 120 and in S. enterica serotype Agona.
Multidrug-resistant Salmonella enterica serotype Typhimurium definitive phage type 104 (DT 104) has emerged during the last decade as a global health problem because of its association with animal and human disease (1). Multidrug-resistant strains of this phage type were first identified from exotic birds in the United Kingdom in the early 1980s and in cattle and humans in the late 1980s but have since become common in other animal species such as poultry, pigs, and sheep. The DT 104 epidemic has now spread worldwide, with several outbreaks since 1996 in the United States and Canada (2–5).
Multidrug-resistant S. Typhimurium DT 104 strains are commonly resistant to ampicillin, chloramphenicol/florfenicol, spectinomycin/streptomycin, sulfonamides, and tetracyclines. The antibiotic-resistance genes are clustered in part of a 43-kb genomic island called Salmonella genomic island I (SGI1), between the thdf and int2 genes of the chromosome (6–10). The int2 gene is part of a retron that has been detected only in serotype Typhimurium (7,8). Downstream of the retron sequence is the yidY gene, which is also found in the chromosome of other S. enterica serotypes (7,8). The antibiotic-resistance gene cluster represents approximately one third of SGI1 and is located at the 3′ end of the structure (7,8). All resistance genes are clustered and are bracketed by two integron structures (Figure 1). The first integron carries the aadA2 gene, which confers resistance to streptomycin and spectinomycin, and a truncated sulI resistance gene. The second contains the beta-lactamase gene blaPSE-1 and a complete sulI gene. Flanked by these two integron structures are the floR gene (6), also called floSt (11) or cmlA-like (9), which confers cross-resistance to chloramphenicol and florfenicol, and the tetracycline-resistance genes tetR and tet (G). Florfenicol resistance and detection of the floR gene by polymerase chain reaction (PCR)-based methods have been proposed as a means for rapidly identifying multidrug-resistant S. Typhimurium DT 104 strains (11), since phage typing is available only in specialized laboratories. However, recently SGI1 has been reported in another phage type of serotype Typhimurium and in serotype Agona, suggesting horizontal transfer of SGI1 (7,10,12). In serotype Agona, SGI1 has the same chromosomal location as in DT 104 strains, except that it lacks the retron sequence found downstream of SGI1; thus it is located between the thdf gene and the yidY gene of the chromosome (7).
Recently, Nastasi and Mammina reported the presence of the floR and tet(G) genes detected by PCR in an S. enterica serotype Paratyphi B strain of biovar Java isolated from a tropical fish in Singapore (13). We examined this isolate to determine the presence of SGI1.
The antibiotic-resistance phenotype of the serotype Paratyphi B strain was assessed by the disk-diffusion assay. All antibiotic disks except for florfenicol were purchased from Bio-Rad (Marnes-La-Coquette, France). Florfenicol disks and the drug itself were obtained from Schering-Plough Animal Health (Kenilworth, NJ). The serotype Paratyphi B strain showed a multidrug-resistance profile typical of serotype Typhimurium DT 104 or serotype Agona strains carrying SGI1, i.e., resistance to ampicillin, chloramphenicol and florfenicol, streptomycin and spectinomycin, sulfonamides, and tetracyclines. Moreover, this strain showed the same resistance level to florfenicol as serotypes Typhimurium DT 104 and Agona, i.e., a florfenicol MIC 32 µg/mL (12). The strain was susceptible to trimethoprim and the quinolones nalidixic acid, enrofloxacin, and ciprofloxacin.
No plasmids were detected in the serotype Paratyphi B strain, suggesting that all antibiotic-resistance genes were chromosomally located. PCR mapping of the typical antibiotic resistance genes and integrons associated with SGI1 was performed as described (Figure 1) (12). PCR amplifications yielded products from genomic DNA extracted from the serotype Paratyphi B strain of the size expected from DNA of serotype Typhimurium DT 104 control strain BN9181 (data not shown) (6,12). Partial nucleotide sequencing of the floR gene showed 100% identity with that of serotype Typhimurium DT 104. Thus, these PCR mapping results indicated that the serotype Paratyphi B strain contains the entire antibiotic-resistance gene cluster of serotype Typhimurium DT 104.
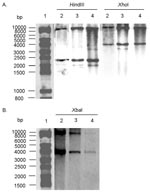
The conservation of the antibiotic-resistance gene’s organization was further assessed by Southern blot of HindIII- or XhoI-digested genomic DNA, with the 12-kb XbaI insert as probe (see XbaI fragment, Figure 1) from recombinant plasmid pSTF3 containing the DT 104 antibiotic-resistance gene cluster as described (6,12). The Southern blot profiles of the serotype Paratyphi B strain were identical to those of the DT 104 control strain BN9181 and serotype Agona strain 959SA97 (Figure 2), confirming that the organization of the antibiotic-resistance genes in the serotype Paratyphi B strain is the same as in serotype Typhimurium DT 104 or serotype Agona containing SGI1 (7,12).
To assess the presence of entire the SGI1 and its location in the chromosome of the serotype Paratyphi B strain, PCR was performed by using primers corresponding to the left and right junctions of SGI1 in the Salmonella chromosome. PCR took into account the presence or absence of the int2-retron sequence, which is located downstream of SGI1 in serotype Typhimurium DT 104 but not in serotype Agona (Figure 1) (7,8). PCR was positive for the left junction of SGI1, as for serotype Typhimurium DT 104 or serotype Agona. If a sequence of the int2 gene of the retron was used as reverse primer, PCR was negative for the right junction of SGI1, but it was positive if the sequence of the yidY gene was used. The PCR products showed the expected sizes of approximately 500 bp for both the left junction and right junction PCR without the retron sequence in Figure 1. These data indicate that the serotype Paratyphi B strain contains SGI1 at the same chromosomal location as in serotype Typhimurium DT 104 or serotype Agona, that is, between the thdf and yidY genes, but lacks the retron sequence found in DT 104 strains and other serotype Typhimurium strains (7,8). The presence of entire SGI1 was also confirmed by Southern blot of XbaI-digested genomic DNA with the p1-9 probe containing a 2-kb EcoRI insert. This corresponds to a central region of SGI1, comprising parts of the S023 and S024 open reading frames, which code for putative helicase and exonuclease proteins, upstream of the antibiotic-resistance gene cluster (7). This probe revealed XbaI fragments of the 4- and 9-kb sizes expected in the serotype Paratyphi B strain and the control serotype Typhimurium DT 104 and serotype Agona strains (Figure 2B).
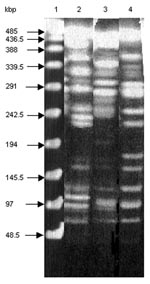
Macrorestriction analysis by pulsed-field gel electrophoresis of the serotype Paratyphi B strain DNA cut by XbaI showed that it is genetically distinct from both serotypes Typhimurium DT 104 and Agona (Figure 3), further indicating at the molecular level that the occurrence of SGI1 in the serotype Paratyphi B strain probably results from horizontal transfer and not seroconversion of known S. enterica serotypes containing SGI1.
We have identified SGI1 in a S. Paratyphi B strain. These data, in conjunction with the identification of SGI1 in S. Agona and S. Typhimurium strains, suggest horizontal transfer of this region (7,10,12). That SGI1 has the same chromosomal location in S. Typhimurium, S. Agona, and S. Paratyphi B suggests that its insertion occurred through a homologous recombination event, perhaps through phage transduction (14). This hypothesis is experimentally supported by the fact that resistance genes of serotype Typhimurium DT 104 can be efficiently transduced by P22-like phage ES18 and phage PDT17, which is released by all DT 104 isolates analyzed (14). However, the question remains why the retron sequence downstream of SGI1 in serotype Typhimurium DT 104 and DT 120 strains (7) is not present downstream of SGI1 in other serotypes. A possible explanation could be that in the horizontal transfers described here, the SGI1 donor strains are not serotype Typhimurium strains. Once SGI1 has been acquired, it may become stable in the chromosome, as in vitro excision experiments failed to demonstrate its loss in a DT 104 strain in the absence of antibiotic selective pressure (8). This factor may contribute to evolution of S. enterica pathogens, similar to the acquisition of pathogenicity islands (15,16).
Acquisition of SGI1 may have been a key factor contributing to the DT 104 worldwide epidemic, perhaps not only through selection by agricultural use of antimicrobial agents (17) but also by virulence properties of SGI1 (10). Therefore, further surveillance is warranted for the emergence of horizontal transfer of SGI to S. enterica serotypes of public health importance.
Dr. Meunier is a postdoctoral fellow at the Institut National de la Recherche Agronomique in France. Her main interests are antibiotic-resistance mechanisms and spread of antibiotic-resistance genes among enterobacterial pathogens.
Acknowledgments
We thank C. Mouline for expert technical assistance.
This work was supported by a grant from the French Institut National de la Recherche Agronomique (INRA, Action Transversalité 2001-2003).
References
- Hancock D, Besser T, Gay J, Rice D, Davis M, Gay C. The global epidemiology of multiresistant Salmonella enterica serovar Typhimurium DT104. In: Brown C, Bolin C, editors. Emerging diseases of animals. Washington: ASM Press; 2000. p. 217-43.
- Besser TE, Goldoft M, Pritchett LC, Khakhria R, Hancock DD, Rice DH, Multiresistant Salmonella Typhimurium DT104 infections of humans and domestic animals in the Pacific Northwest of the United States. Epidemiol Infect. 2000;124:193–200. DOIPubMedGoogle Scholar
- Davis MA, Hancock DD, Besser TE, Rice DH, Gay JM, Gay C, Changes in antimicrobial resistance among Salmonella enterica serovar Typhimurium isolates from humans and cattle in the Northwestern United States, 1982-1997. Emerg Infect Dis. 1999;5:802–6.PubMedGoogle Scholar
- Glynn MK, Bopp C, Dewitt W, Dabney P, Mokhtar M, Angulo FJ. Emergence of multidrug-resistant Salmonella enterica serotype Typhimurium DT104 infections in the United States. N Engl J Med. 1998;338:1333–8. DOIPubMedGoogle Scholar
- Poppe C, Smart N, Khakhria R, Johnson W, Spika J, Prescott J. Salmonella typhimurium DT104: a virulent and drug-resistant pathogen. Can Vet J. 1998;39:559–65.PubMedGoogle Scholar
- Arcangioli MA, Leroy-Sétrin S, Martel JL, Chaslus-Dancla E. A new chloramphenicol and florfenicol resistance gene flanked by two integron structures in Salmonella typhimurium DT104. FEMS Microbiol Lett. 1999;174:327–32. DOIPubMedGoogle Scholar
- Boyd D, Peters GA, Cloeckaert A, Sidi Boumedine K, Chaslus-Dancla E, Imberechts H, Complete nucleotide sequence of a 43-kilobase genomic island associated with the multidrug resistance region of Salmonella enterica serovar Typhimurium DT104 and its identification in phage type DT120 and serovar Agona. J Bacteriol. 2001;183:5725–32. DOIPubMedGoogle Scholar
- Boyd DA, Peters GA, Ng LK, Mulvey MR. Partial characterization of a genomic island associated with the multidrug resistance region of Salmonella enterica Typhimurium DT104. FEMS Microbiol Lett. 2000;189:285–91. DOIPubMedGoogle Scholar
- Briggs CE, Fratamico PM. Molecular characterization of an antibiotic resistance gene cluster of Salmonella typhimurium DT104. Antimicrob Agents Chemother. 1999;43:846–9.PubMedGoogle Scholar
- Cloeckaert A, Schwarz S. Molecular characterization, spread and evolution of multidrug resistance in Salmonella enterica Typhimurium DT104. Vet Res. 2001;32:301–10. DOIPubMedGoogle Scholar
- Bolton LF, Kelley LC, Lee MD, Fedorka-Cray PJ, Maurer JJ. Detection of multi-drug resistant Salmonella enterica serotype typhimurium DT104 based on a gene which confers cross-resistance to florfenicol and chloramphenicol. J Clin Microbiol. 1999;37:1348–51.PubMedGoogle Scholar
- Cloeckaert A, Sidi Boumedine K, Flaujac G, Imberechts H, D’Hooghe I, Chaslus-Dancla E. Occurrence of a Salmonella enterica serovar Typhimurium DT104-like antibiotic resistance gene cluster including the floR gene in S. enterica serovar Agona. Antimicrob Agents Chemother. 2000;44:1359–61. DOIPubMedGoogle Scholar
- Nastasi A, Mammina C. Presence of class I integrons in multi-drug resistant, low prevalence Salmonella serotypes, Italy. Emerg Infect Dis. 2001;7:455–7.PubMedGoogle Scholar
- Schmieger H, Schicklmaier P. Transduction of multiple drug resistance of Salmonella enterica serovar Typhimurium DT104. FEMS Microbiol Lett. 1999;170:251–6. DOIPubMedGoogle Scholar
- Hansen-Wester I, Hensel M. Salmonella pathogenicity islands encoding type III secretion systems. Microbes Infect. 2001;3:549–59. DOIPubMedGoogle Scholar
- Hentschel U, Hacker J. Pathogenicity islands: the tip of the iceberg. Microbes Infect. 2001;3:545–8. DOIPubMedGoogle Scholar
- Angulo FJ, Griffin PM. Changes in antimicrobial resistance in Salmonella enterica serovar Typhimurium. Emerg Infect Dis. 2000;6:436–7.PubMedGoogle Scholar
Figures
Cite This ArticleTable of Contents – Volume 8, Number 4—April 2002
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Axel Cloeckaert, Unité de Pathologie Aviaire et Parasitologie, Institut National de la Recherche Agronomique, 37380 Nouzilly, France; fax: 33- 2 47 42 77 74;
Top