Volume 11, Number 1—January 2005
Research
Human Parechovirus-3 and Neonatal Infections
Figure 2
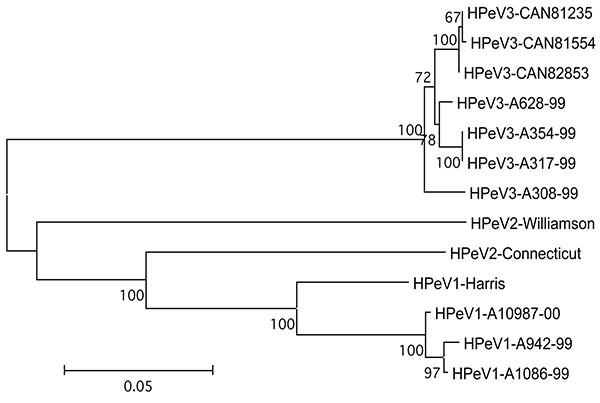
Figure 2. Phylogenetic tree showing the relationship between human parechovirus (HPeV)-3 Canadian isolates no. 81235, 81554, and 82853 and other HPeV-3 (A628-99, GenBank accession no. AB112484; A354-99, no. AB112483; A317-99, no. AB112482; A308-99, no. AB084913), HPeV-2 (Williamson, no. AJ005695; Connecticut, no. AF055846) and HPeV-1 (Harris, no. S45208; A10987-00, no. AB112487; A942-99, no. AB112486; A1086-99, no. AB112485) strains based on amino acid differences in capsid proteins (VP0-VP3-VP1 region). The tree was constructed by using the neighbor-joining method. Numbers represent the frequency of occurrence of nodes in 500 bootstrap replicas.
Page created: June 16, 2011
Page updated: June 16, 2011
Page reviewed: June 16, 2011
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.