Volume 13, Number 8—August 2007
Dispatch
Pathogenic Hantaviruses, Northeastern Argentina and Eastern Paraguay
Figure 2
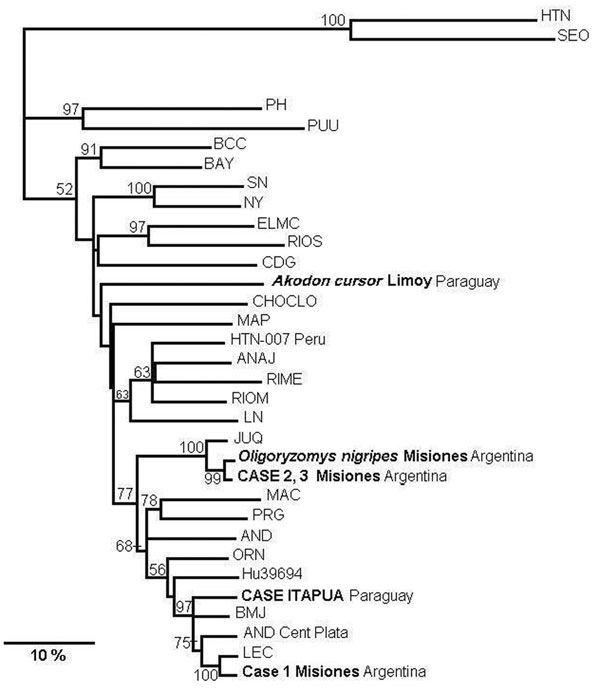
Figure 2. Phylogenetic relationships among the nucleotide sequences of the N protein of different hantaviruses from North America. A maximum parsimonious phylogenetic tree was generated on the basis of nucleotide sequence differences in the 904-nt region of the N gene open reading frame, which is available for South American strains by using PHYLIP version 3.57c. Bootstrap values >50%, obtained from 1,000 replicates of the analysis, are shown for the branch points. Lengths of the horizontal branches are proportional to the nucleotide step differences. The strain sequences under study in this paper are in italics. The following published S-segment sequences were included in the analysis (GenBank accession no.): Hantaan (HTN; U37768), Seoul (SEO; AB027522), Prospect Hill (PH; Z49098), Puumala (PUU; X61035), Black Creek Canal (BCC; L39949), Bayou (BAY; L36929), Sin Nombre (SN; L25784), New York (NY; U36801), El Moro Canyon (ELMC; U11427), Río Segundo (RIOS; U18100), Caño Delgadito (CDG; AF000140), Choclo (CHOCLO; DO285046), Maporal (MAP; AY267347), HTN-007 Perú (HTN-007 Perú. AF133254), Anajatuba (ANAJ; DQ451829), Rio Mearim (RIME; DQ451828), Río Mamoré Bolivia (RIOM; U52136), Laguna Negra (LN; AF005727), Araucaria (JUQ; AY740633), Maciel (MAC; AF0482716), Pergamino (PRN; 482717), Andes (AND; AF324902), Oran (ORN; AF028024), Hu39694 (Hu39694; AF482711), Bermejo (BMJ; AF482713), Lechiguanas (LEC; AF482714).