Volume 15, Number 5—May 2009
Research
New Respiratory Enterovirus and Recombinant Rhinoviruses among Circulating Picornaviruses
Figure 2
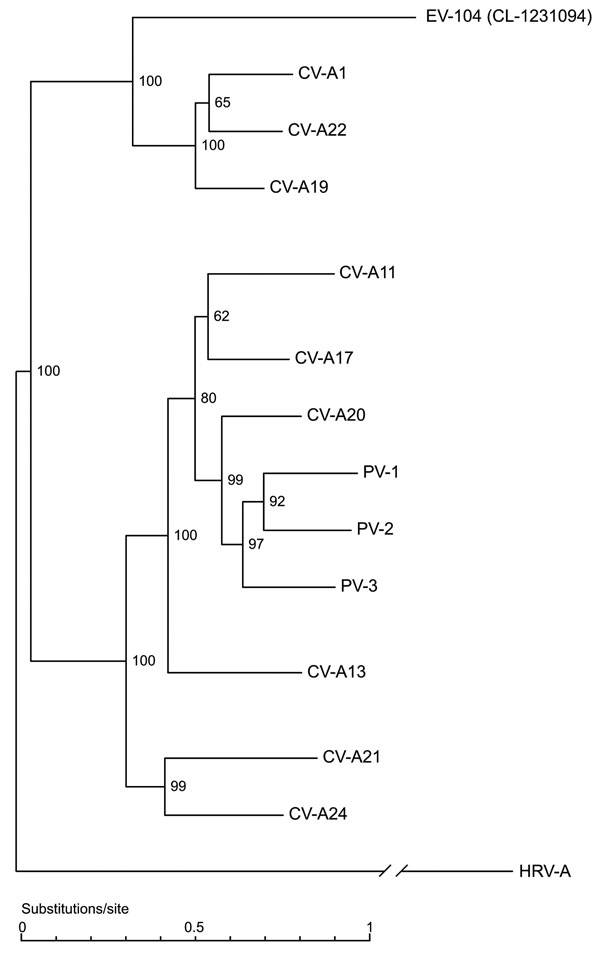
Figure 2. Full genome phylogenetic tree of enterovirus 104 (EV-104), representative strain CL-1231094, and members of the human enterovirus C (HEV-C) species. Human rhinovirus A (HRV-A) (GenBank accession no. DQ473509) was used as outgroup. Coxsackievirus A1 (CV-A1) (AF499635), CV-A21 (AF546702), CV-A20 (AF499642), CV-A17 (AF499639), CV-A13 (AF499637), CV-A11 (AF499636), CV-A19 (AF499641), CV-A22 (AF499643), CV-A24 (D90457), poliovirus 1 (PV-1) (V01148), PV-2 (X00595), and PV-3 (X00925) sequences were obtained from GenBank.
Page created: December 16, 2010
Page updated: December 16, 2010
Page reviewed: December 16, 2010
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.