Volume 16, Number 3—March 2010
Dispatch
Rare Influenza A (H3N2) Variants with Reduced Sensitivity to Antiviral Drugs
Figure 2
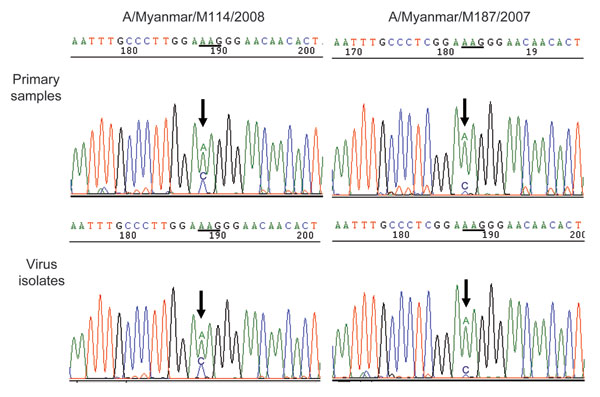
Figure 2. Detection of Q136K substitution in NA by sequencing in primary samples and virus isolates. Arrows indicate the first peak of the codon encoding amino acid position 136. Comparison of the sequence chromatogram showed a mixed population of bases in both original clinical samples and virus isolates, with a dominant peak for 136K (AAG) mutants, compared with wild-type 136Q (CAG) viruses. NA, neuraminidase.
Page created: December 14, 2010
Page updated: December 14, 2010
Page reviewed: December 14, 2010
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.