Volume 12, Number 11—November 2006
Dispatch
Reemergence of Dengue Virus Type 4, French Antilles and French Guiana, 2004–2005
Abstract
After 10 years of absence, dengue virus type 4 (DENV-4) has recently reemerged in Martinique, Guadeloupe, and French Guiana. Phylogenetic analyses of strains isolated from 2004 to 2005 showed that they belong to DENV-4 genotype II, but to a different cluster than strains isolated from 1993 to 1995.
Dengue is a viral disease transmitted by mosquitoes. It is caused by any of the 4 viral serotypes of dengue virus (DENV), designated DENV-1, DENV-2, DENV-3, and DENV-4. DENV belongs to the genus Flavivirus and family Flaviviridae. Flaviviruses are enveloped, single-stranded, positive-sense RNA viruses. The genomic RNA is ≈11 kb and has 10 genes coding 3 structural proteins (capsid [C], envelope [E], and membrane [M]), and 7 nonstructural proteins (NS1, NS2a, NS2b, NS3, NS4a, NS4b, and NS5).
Dengue is the predominant arthropodborne viral disease affecting humans. DENV causes a wide range of symptoms from inapparent or mild disease (dengue fever) to severe forms such as dengue hemorrhagic fever (DHF) and dengue shock syndrome (DSS). The disease is now endemic in >100 countries and threatens >2.5 billion people. It currently occurs in tropical areas and affects <100 million persons each year (1). Of these persons, 500,000 have DHF and ≈25,000, mainly children, die. The World Health Organization has estimated a 30-fold incidence increase in dengue the past 50 years (2).
DENV is transmitted to humans by Stegomyia aegypti (formerly Aedes aegypti) mosquitoes. Before the 1980s, epidemic dengue was rare in the Americas because St. aegypti had been eradicated from most Central and South American countries. In the 1990s, St. aegypti had almost completely reinvaded the regions in which it was found before its eradication. Within the past 30 years, the increase in the worldwide transportation network, as well as uncontrolled population growth and urbanization, has led to larger and more frequent DENV epidemics and more cases of DHF/DSS (3).
During the past 20 years, the 4 DENV serotypes have been isolated in the French departments of the Americas: Guadeloupe, Martinique, and French Guiana. Martinique and Guadeloupe are 2 Caribbean islands located in the Lesser Antilles and represent the French West Indies, and French Guiana is located southeast of the French West Indies in northern South America between Suriname and Brazil. DENV is endemic in French Guiana. Dengue epidemics occurred in this country at 4–6-year intervals from the 1960s to the early 1990s (4). The first DHF cases were associated with a DENV-2 outbreak in 1991–1992 (5). DENV-4 was sporadically isolated between 1993 and 1995. Cocirculation of DENV-1 and DENV-2 caused outbreaks between 1996 and 1998, followed by 2 consecutive DENV-3 epidemics in 2001–2002 and 2004–2005. Martinique had 3 dengue epidemics during the past decade in a setting of sporadic transmission and seasonal peaks from July to December. These epidemics were associated with DENV-2 and DENV-4 in 1995, DENV-1 in 1997, and DENV-3 in 2001 (6). The epidemiology of DENV in Guadeloupe is similar to that in Martinique, although no data are available about previous dengue epidemics.
Our study reports the evolution of DENV-4 in French Guiana and the French Caribbean islands since its last detection and circulation from 1993 to 1995. We studied 8 DENV-4 strains isolated from human sera from French Guiana in 1993 and 1995 (n = 6) and in 2004 and 2005 (n = 2) (Table). We also tested 3 human serum specimens from Martinique (n = 2) and Guadeloupe (n = 1) that were positive for DENV-4 during dengue surveillance in the fourth quarter of 2004.
DENV-4 infection was confirmed by using virus isolation on AP 61 cells as previously described (7). We then amplified from first-passage RNA a 1,940-bp region of the genome for the E gene and adjacent prM/M and NS1 junctions previously described (8) to analyze the phylogenetic relationships of these strains. Each PCR product was cloned by using the TOPO TA Cloning kit (Invitrogen, Paisley, UK). For each isolate, 3 clones were sequenced by Genome Express (Meylan, France).
We compared our 11 sequences to the 87 DENV-4 sequences available in the GenBank database. Sequences were aligned by using MacVector version 7.2 software (Oxford Molecular Ltd., Oxford, UK). Percentage of nucleotide identity between sequences was determined for all observed differences (including insertions and deletions) and was calculated for the 1,473 nt available for all 98 sequences analyzed. Sequence comparison of strains from Martinique, Guadeloupe, and French Guiana with those reported from Asia showed that nucleotide divergence is 8%–12%. The level of nucleotide divergence was lower for sequences from the Caribbean area (maximum nucleotide divergence 2.8%). All 11 newly obtained sequences were different from each other. These sequences exhibited a 0.06%–0.47% nucleotide divergence (strains isolated in 1993 and 1995) and a 0.06%–1.22% nucleotide divergence (strains isolated in 2004 and 2005) among themselves. These strains had amino acid changes at position 351 in the E gene (from isoleucine to valine) and position 52 in the NS1 gene (from lysine to arginine), which supports their inclusion in the modern Caribbean basin clade (8).
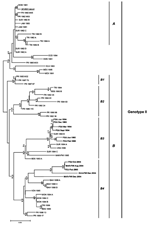
We conducted phylogenetic analyses on nucleotide sequences by using distance and parsimony methods in PAUP* version 4.0b8 to increase the reliability of derived tree topologies (9). We also used the neighbor-joining distance matrix algorithm with the Kimura 2 parameter (Figure) and the heuristic algorithm for the maximum parsimony analysis (data not shown). Robustness of resulting topologies was examined by using bootstrap analyses (10). Both neighbor-joining and parsimony algorithms underwent 1,000 iterations.
The phylogenetic tree constructed with the neighbor-joining method on nucleotide sequences restricted to all American DENV-4 isolates available in GenBank (Table) is shown in the Figure. Our 11 strains segregate into 2 main clades that were identified in the 2 phylogenetic analyses from consistent topologic associations and high bootstrap values. The strains isolated from French Guiana in 1993 and 1994 clustered with strains from Suriname and Costa Rica, which were isolated at the same time, as well as with a previously reported strain from Martinique isolated in 1995. Our 5 strains isolated in 2004 and 2005 are related to a group of DENV-4 genotype II sequences isolated in the Bahamas in 1998. This finding suggests that strains circulating in French departments of the Americas in 2004 and 2005 probably evolved from strains detected in the Caribbean islands during the late 1990s. Our observations confirm that DENV-4 lineages in the Caribbean and nearby regions are grouped temporally rather than by the geographic origin of the isolates, as has been previously suggested (8).
The DEN-4 serotype was first reported in the Americas in 1981. At that time, isolates collected in the Americas were similar to strains collected in Southeast Asia. This lineage was designated DENV-4 genotype II (11). Since it was first detected in Dominica and the French islands of Saint Barthelemy and Saint Martin (12), this genotype has spread rapidly throughout the Caribbean and Latin America and caused DF and sporadic cases of DHF/DSS (3).
DENV-4 genotype II never stopped circulating in the Caribbean region, despite its being periodically locally eliminated (8). This genotype reappeared in French departments of the Americas in 2004 after the introduction of a strain circulating in neighboring Caribbean islands, rather than by reemergence of a strain detected 10 years ago. Furthermore, this DENV-4 genotype was detected in French Guiana (October 2004) a few weeks after it was detected in Martinique (August 2004), which suggests that the strain came from the Antilles (Table). A similar phenomenon had already been observed in 1999 for the DENV-3 genotype (J. Morvan, unpub. data). This model of viral spread may be caused by cultural and economic ties between French departments, which allows gene flow through the Caribbean islands and South America as persons with viremia move through the region. This reflects the general pattern of dengue evolution in the Americas (13,14).
Reintroduction of DENV-4 in the French Antilles and French Guiana in 2004 after a decade of absence highlights the risk for subsequent epidemics in persons with no immunity to dengue. As expected, the French Antilles had a major DENV-4 epidemic with cocirculation of DENV-2 from July 2005 to January 2006 (15). In contrast, only sporadic cases of DENV-4 infection have been observed in French Guiana since the beginning of 2005. This indicates that other factors such as mosquito density or human susceptibility, in addition to immune status of the host or silent transmission of the virus, can modulate the risk for dengue epidemics.
Dr Dussart is a virologist at the Laboratoire de Virologie, Institut Pasteur de la Guyane. He is also in charge of the Centre National de Réference des Arbovirus et Virus Influenza region Antilles Guyane. His research interests include diagnosis and epidemiology of dengue virus and other arboviruses.
Acknowledgment
We thank Jacques Rosine, Pascal Chaud, Thierry Cardoso, Alain Blateau, and Philippe Quenel from the CIRE Antilles-Guyuane for their helpful support. Sequences reported in this paper have been deposited in the GenBank database (accession nos. DQ390319–DQ390329).
References
- World Health Organization. Epidemic and pandemic alert and response: impact of dengue. [cited 2006 Sep 18]. Available from http://www.who.int/csr/disease/dengue/impact/en/index.html
- Gubler DJ. Dengue and dengue hemorrhagic fever. Clin Microbiol Rev. 1998;11:480–96.PubMedGoogle Scholar
- Fouque F, Reynes JM, Moreau JP. Dengue in French Guiana, 1969–1993. Bull Pan Am Health Organ. 1995;29:147–55.PubMedGoogle Scholar
- Reynes JM, Laurent A, Deubel V, Telliam E, Moreau JP. The first epidemic of dengue hemorrhagic fever in French Guiana. Am J Trop Med Hyg. 1994;51:545–53.PubMedGoogle Scholar
- Peyrefitte CN, Couissinier-Paris P, Mercier-Perennec V, Bessaud M, Martial J, Kenane N, Genetic characterization of newly reintroduced dengue virus type 3 in Martinique (French West Indies). J Clin Microbiol. 2003;41:5195–8. DOIPubMedGoogle Scholar
- Tran A, Deparis X, Dussart P, Morvan J, Rabarison P, Remy F, Dengue spatial and temporal patterns, French Guiana, 2001. Emerg Infect Dis. 2004;10:615–21.PubMedGoogle Scholar
- Foster JE, Bennett SN, Vaughan H, Vorndam V, McMillan WO, Carrington CV. Molecular evolution and phylogeny of dengue type 4 virus in the Caribbean. Virology. 2003;306:126–34. DOIPubMedGoogle Scholar
- Swofford DL. (1998). PAUP*: phylogenetic analysis using parsimony (*and other methods), Version 4.0b8. Sunderland (MA): Sinauer Associates.
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–91. DOIGoogle Scholar
- Lanciotti RS, Gubler DJ, Trent DW. Molecular evolution and phylogeny of dengue-4 viruses. J Gen Virol. 1997;78:2279–86.PubMedGoogle Scholar
- Centers for Disease Control and Prevention. Dengue type 4 infections in U.S. travelers to the Caribbean. MMWR Morb Mortal Wkly Rep. 1981;30:249–50.PubMedGoogle Scholar
- Foster JE, Bennett SN, Carrington CV, Vaughan H, McMillan WO. Phylogeography and molecular evolution of dengue 2 in the Caribbean basin, 1981–2000. Virology. 2004;324:48–59. DOIPubMedGoogle Scholar
- Carrington CV, Foster JE, Pybus OG, Bennett SN, Holmes EC. Invasion and maintenance of dengue virus type 2 and type 4 in the Americas. J Virol. 2005;79:14680–7. DOIPubMedGoogle Scholar
- Bulletin d'alertes et de surveillance Antilles Guyane. Epidémiologie de dengue en Martinique et en Guadeloupe. Numéro spécial: suivi des épidémies de dengue aux Antilles Guyane [cited 2005 Nov]. Available from http://www.martinique.sante.gouv.fr/documents/accueil/cire/Basag2005-13.pdf
Figure
Table
Cite This ArticleTable of Contents – Volume 12, Number 11—November 2006
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Philippe Dussart, Centre National de Référence des Arbovirus, Institut Pasteur de la Guyane, 23 Ave Pasteur, BP 6010, 97306 Cayenne CEDEX, French Guiana
Top