Volume 13, Number 9—September 2007
Dispatch
Hantavirus in Northern Short-tailed Shrew, United States
Figure
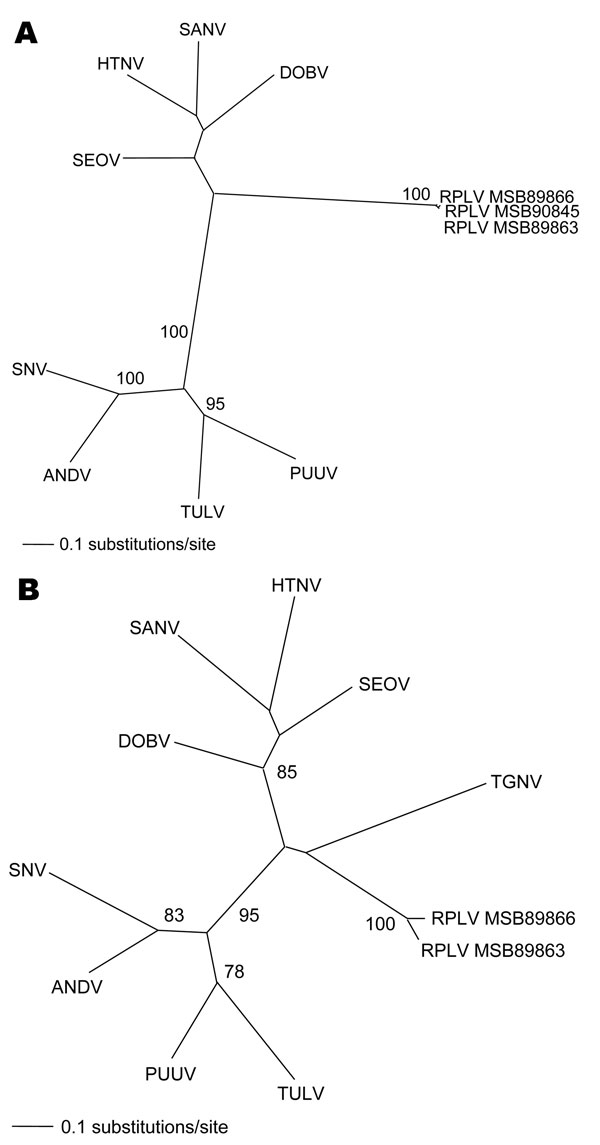
Figure. Phylogenetic trees generated by maximum likelihood method and generalized time reversible + I + G model of evolution as estimated from data on alignment of the partial (A) 1,390-nt medium (M)– and (B) 490-nt large (L)–genomic segments of Camp Ripley virus (RPLV). Phylogenetic positions of RPLV strains MSB89863, MSB89866, and MSB90845 are shown in relationship to representative Murinae rodentborne hantaviruses, including Hantaan virus (HTNV 76–118, NC_005219, NC_005222), Sangassou virus (SANV SA14, DQ268651, DQ268652), Dobrava virus (DOBV AP99, NC_005234, NC_005235), and Seoul virus (SEOV 80 39, NC_005237, NC_005238); Arvicolinae rodentborne hantaviruses, including Tula virus (TULV M5302v, NC_005228, NC_005226) and Puumala virus (PUUV Sotkamo, NC_005223, NC_005225); and Neotominae and Sigmodontinae rodentborne hantaviruses, including Andes virus (ANDV Chile 9717869, NC_003467, NC_003468) and Sin Nombre virus (SNV NMH10, NC_005215, NC_005217). Tanganya virus (TGNV Tan826, EF050454) from the Therese shrew (Crocidura theresae) is also shown. Host identification of Blarina brevicauda was confirmed by morphologic assessment of voucher specimens and by mitochondrial DNA sequences (data not shown). The numbers at each node are bootstrap support values (expressed as the percentage of replicates in which the node was recovered), as determined for 100 maximum likelihood iterations under the same model of evolution by PAUP version 4.0 (http://paup.csit.fsu.edu). The scale bar indicates 0.1 nt substitutions per site. GenBank accession nos. for RPLV: M (EF540774, EF540775, EF540773) and L (EF540771, EF540772).