Volume 14, Number 11—November 2008
Dispatch
Unusual Cryptosporidium Genotypes in Human Cases of Diarrhea
Abstract
Several Cryptosporidium spp. are known to infect humans, but most cases of illness are caused by Cryptosporidium hominis or C. parvum. During a long-term genotyping in the United Kingdom, we identified 3 unusual Cryptosporidium genotypes (skunk, horse, and rabbit) in human patients with diarrhea.
Cryptosporidium spp. are frequently a cause of diarrheal disease in immunocompetent as well as immunocompromised humans. Over the past decade molecular methods have enabled the characterization and identification of species and genotypes within the genus. The taxonomy is under continual review, but so far 20 valid species and numerous genotypes have been described. Many are named after the original host from which the isolate was recovered and are often referred to as “host-adapted” (1,2). Most human infections are caused by C. hominis or C. parvum but C. meleagridis, C. felis, C. canis, C. suis, C. muris, C. andersoni, C. hominis monkey genotype, cervine genotype, and the chipmunk genotype I have also been detected (1–6). The immune status of the host is not necessarily linked to infection with other species/genotypes (1,7). We describe 3 unusual Cryptosporidium genotypes detected in human patients with diarrhea.
Since 2000, the UK Cryptosporidium Reference Unit has maintained a national collection of Cryptosporidium oocysts (8). Over 16,000 Cryptosporidium-positive human fecal samples have been submitted by primary diagnostic laboratories and characterized by the Reference Unit to identify the infecting species. In addition to the expected C. hominis, C. parvum, and small number of C. meleagridis, C. felis, C. canis, and cervine genotype isolates, 3 other genotypes (skunk, horse, and rabbit) were identified in separate samples from individual patients after the onset of diarrhea in 2000 (sample W971), 2003 (sample W6863), and 2007 (sample W16103). A routinely collected minimum dataset was submitted with each sample, and further exposure data were collected for each patient from the local Consultant in Communicable Disease Control.
To prepare isolates for molecular characterization, oocysts were concentrated by saturated salt flotation, disrupted by boiling for 1 hour and the DNA purified by using a QIAamp DNA Mini Kit (QIAGEN, Valencia, CA, USA) as previously described (9). All 3 isolates were characterized by PCR–restriction fragment length polymorphism (RFLP) or bidirectional sequencing (GeneService Ltd., Cambridge, UK) at the small subunit (SSU) rRNA (≈800-bp product) (10), Cryptosporidium oocyst wall protein (COWP) (≈550-bp product) (11) and heat shock protein (HSP) 70 (≈450-bp or ≈325-bp products) (12) genes. Sequences were compared with GenBank submissions by using the BLAST algorithm (www.ncbi.nlm.nih.gov/Education/BLASTinfo/BLAST_algorithm.html).
To confirm identification, phylogenetic analysis was conducted in TREECON (www.bioinformatics.psb.ugent.be/software/details/3) with other known Cryptosporidium spp. and genotypes by using alignments generated in ClustalX version 2.0 (ftp://ftp.ebi.ac.uk/pub/software/clustalw2) and manually edited in BioEdit version 7.0.9 (www.mbio.ncsu.edu/BioEdit/bioedit.html). All sequences generated in this study have been submitted to GenBank under accession nos. EU437411–EU437418.
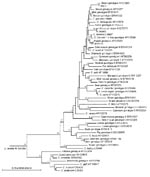
At the SSU rRNA and HSP70 genes, sequence analysis confirmed that W971, W6863, and W16103 were skunk, horse, and rabbit genotypes, respectively (Table). Isolate W971 was homologous with genotype W13 found in storm water, which, in turn, is the skunk genotype (6). Initially, the BLAST search for isolate W6863 erroneously indicated C. parvum as the most probable identity at the SSU rRNA gene, but this was due to the short length (484 bp) of the only horse genotype sequence available (AY273770) for comparison. Thus, C. parvum isolates that spanned our whole query sequence (787 bp) were calculated to have greater identities by BLAST. However, a detailed comparison between AY273770 and W6863 showed only 2-bp differences (including 1 insertion in our sequence) compared with 7-bp differences between W6863 and C. parvum. W6863 was confirmed as a variant of the horse genotype by HSP70 gene sequence analysis and SSU rRNA gene phylogenetic analysis (Figure).
PCR-RFLP analysis of the SSU rRNA and COWP genes differentiated the skunk and horse genotypes from the most common human pathogens. However, identifying the rabbit genotype by PCR-RFLP at these loci was more problematic because of this genotype’s close relationship with C. hominis. The sequence and restriction pattern are identical at the COWP gene and, with only 4-bp substitutions (2 occurring in SspI cut-sites), the pattern is similar at the SSU rRNA gene. Increasing the resolution by running the agarose gel at an appropriate concentration and for as long as possible is important for the separation of the C. hominis diagnostic band (449 bp) from the rabbit genotype (472 bp).
Information on possible risk factors was collected for the 2 weeks before the onset of illness, but we cannot be sure how these 3 persons became infected with the unusual genotypes. The skunk genotype was found in a 25-year-old woman from a rural area of southwest England, who reported no foreign travel and no contact with animals. She worked and swam regularly at an adult daycare center and had spent a week during the incubation period with clients at a holiday forest park in her region. There was no information to suggest that she was immunocompromised. The horse genotype was found in a 30-year-old immunocompetent woman also from a rural area of southwest England, who reported swimming and foreign travel (destination unknown) but no contact with animals during the incubation period. The rabbit genotype was found in a 48-year-old immunocompetent woman from a rural area of northwest England, who reported foreign travel to southern Spain and contact with wild birds (feeding ducks and geese) but no contact with other animals.
Previously, these 3 genotypes were known to cause infections only in wild or zoo animals (13,14). Wild animals are known to be an important source of Cryptosporidium oocysts in environmental samples and we have detected the rabbit genotype in surface waters and septic tank samples (unpub. data), but the source is unknown. Since many isolates have yet to be found in humans and although little is actually known about them, they are assumed to be insignificant to public health (6,15). The importance of unusual genotypes in humans who seek treatment for diarrheal disease warrants further investigation.
Dr Robinson is a parasitologist at the UK Cryptosporidium Reference Unit, National Public Health Service for Wales. His main research interests include molecular epidemiology, veterinary/medical parasitology, and entomology.
Acknowledgments
We thank the consultants at the Communicable Disease Control and Health Protection Units for providing additional patient information; the staff at the UK Cryptosporidium Reference Unit for scientific, administrative, and technical support; the primary diagnostic laboratories for contributing the specimens; and Meirion Evans for his helpful comments.
Samples were collected as part of the National Collection of Cryptosporidium Oocysts, in part funded by Welsh Assembly Government and Department for Environment, Food & Rural Affairs (administered by the Drinking Water Inspectorate).
References
- Xiao L, Fayer R, Ryan U, Upton SJ. Cryptosporidium taxonomy: recent advances and implications for public health. Clin Microbiol Rev. 2004;17:72–97. DOIPubMedGoogle Scholar
- Xiao L, Ryan U. Cryptosporidiosis: an update in molecular epidemiology. Curr Opin Infect Dis. 2004;17:483–90. DOIPubMedGoogle Scholar
- Morse TD, Nichols RA, Grimason AM, Campbell BM, Tembo KC, Smith HV. Incidence of cryptosporidiosis species in paediatric patients in Malawi. Epidemiol Infect. 2007;135:1307–15. DOIPubMedGoogle Scholar
- Leoni F, Amar C, Nichols G, Pedraza-Diaz S, McLauchlin J. Genetic analysis of Cryptosporidium from 2,414 humans with diarrhoea in England between 1985 and 2000. J Med Microbiol. 2006;55:703–7. DOIPubMedGoogle Scholar
- Feltus DC, Giddings CW, Schneck BL, Monson T, Warshauer D, Mcevoy JM. Evidence supporting zoonotic transmission of Cryptosporidium spp. in Wisconsin. J Clin Microbiol. 2006;44:4303–8. DOIPubMedGoogle Scholar
- Feng Y, Alderisio KA, Yang W, Blancero LA, Kuhne WG, Nadareski CA, Cryptosporidium genotypes in wildlife from a New York watershed. Appl Environ Microbiol. 2007;73:6475–83. DOIPubMedGoogle Scholar
- Chalmers RM, Elwin K, Thomas A, Joynson DHM. Infection with unusual types of Cryptosporidium is not restricted to immunocompromised patients. J Infect Dis. 2002;185:270–1. DOIPubMedGoogle Scholar
- The Development of a National Collection for Oocysts of. Cryptosporidium. DWI 170/2/125. Marlow (UK): Foundation for Water Research; 2002.
- Elwin K, Chalmers RM, Roberts R, Guy EC, Casemore DP. Modification of a rapid method for the identification of gene-specific polymorphisms in Cryptosporidium parvum and its application to clinical and epidemiological investigations. Appl Environ Microbiol. 2001;67:5581–4. DOIPubMedGoogle Scholar
- Xiao L, Alderisio K, Limor J, Royer M, Lal AA. Identification of species and sources of Cryptosporidium oocysts in storm waters with a small-subunit rRNA-based diagnostic and genotyping tool. Appl Environ Microbiol. 2000;66:5492–8. DOIPubMedGoogle Scholar
- Spano F, Putignani L, McLauchlin J, Casemore DP, Crisanti A. PCR-RFLP analysis of the Cryptosporidium oocyst wall protein (COWP) gene discriminates between C. wrairi and C. parvum, and between C. parvum isolates of human and animal origin. FEMS Microbiol Lett. 1997;150:209–17.PubMedGoogle Scholar
- Morgan UM, Monis PT, Xiao L, Limor J, Sulaiman I, Raidal S, Molecular and phylogenetic characterisation of Cryptosporidium from birds. Int J Parasitol. 2001;31:289–96. DOIPubMedGoogle Scholar
- Xiao L, Sulaiman IM, Ryan UM, Zhou L, Atwill ER, Tischler ML, Host adaptation and host-parasite co-evolution in Cryptosporidium: implications for taxonomy and public health. Int J Parasitol. 2002;32:1773–85. DOIPubMedGoogle Scholar
- Ryan U, Xiao L, Read C, Zhou L, Lal AA, Pavlasek I. Identification of novel Cryptosporidium genotypes from the Czech Republic. Appl Environ Microbiol. 2003;69:4302–7. DOIPubMedGoogle Scholar
- Zhou L, Fayer R, Trout JM, Ryan UM, Schaefer FW III, Xiao L. Genotypes of Cryptosporidium species infecting fur-bearing mammals differ from those of species infecting humans. Appl Environ Microbiol. 2004;70:7574–7. DOIPubMedGoogle Scholar
Figure
Table
Cite This ArticleTable of Contents – Volume 14, Number 11—November 2008
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Rachel M. Chalmers, UK Cryptosporidium Reference Unit, National Public Health Service Microbiology Swansea Singleton Hospital, Swansea, SA2 8QA, UK;
Top