Volume 14, Number 3—March 2008
Research
Genetic Variability of West Nile Virus in US Blood Donors, 2002–2005
Figure 3
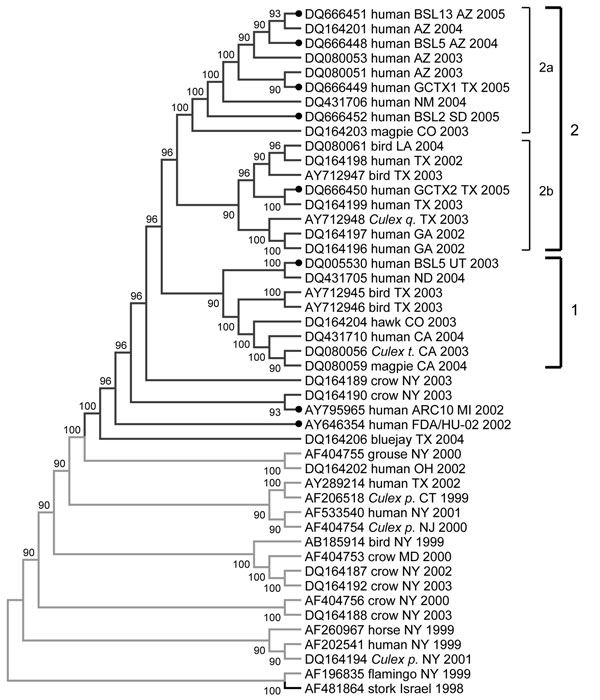
Figure 3. Phylogenetic tree of complete genomes of West Nile virus (WNV) isolates collected during the 1999–2005 epidemics in the United States. Phylogeny reconstruction was estimated by using MEGA version 3.1 (www.megasoftware.com) on the basis of maximum parsimony analysis. Solid circles indicate isolates from this study. Values near branches represent percentage support by parsimony bootstrap analysis. Some parsimony-informative positions (1442, 2446, 4146, 6138, 6721, 8811, 10408, and 10851) play an important role in topologic arrangement of the tree and outgroup configurations. The tree was rooted with prototype WNV isolate WN-NY99 (AF196835) and the most closely related Old World isolate, IS-98 (AF481864). Culex q., Culex. quinquefasciatus; Culex t., Cx. tarsalis; Culex p., Cx. pipiens. WNV genotype is color coded: green, WN99; blue, WN02.