Volume 19, Number 1—January 2013
Letter
Cronobacter sakazakii ST4 Strains and Neonatal Meningitis, United States
To the Editor: To overcome various limitations of phenotyping and 16S rDNA sequence analysis of Cronobacter bacteria, we have established a comprehensive multilocus sequence typing (MLST) scheme as an open access database resource (www.pubMLST.org/cronobacter) (1). The scheme is based on 7 housekeeping genes (atpD, fusA, glnS, gltB, gyrB, infB, ppsA; 3,036 nt concatenated length) and has been used to study the diversity of the Cronobacter genus and new Cronobacter species (2–4). Previously, we compared the sequence type profile to severity of infection by compiling patient details, isolation site, and clinical signs and symptoms for strains isolated from around the world during 1953–2008 (5). This study revealed that most serious meningitis clinical cases caused by Cronobacter spp. in neonates during the previous 30 years in 6 countries were caused by a single sequence type (ST): C. sakazakii ST4. We were therefore interested in applying the MLST method to the Cronobacter strains associated with the highly publicized cases in the United States during December 2011 (6).
The Centers for Disease Control and Prevention (CDC) sent us the Cronobacter isolates they collected during 2011 for MLST analysis (Table). Ten specimens were clinical isolates from neonates or infants. These included 2 specimens (1577, 1579) associated with Cronobacter infections in Missouri and Illinois (6). Four specimens were from opened tins of powdered infant formula (PIF), and 1 was from PIF reconstitution water. DNA sequences for all specimens are available for download and independent analysis through the open access database.
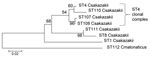
Most (14/15) specimens were C. sakazakii; 1 was C. malonaticus. This predominance of C. sakazakii isolates matches reports of cases and outbreak studies (7). The C. sakazakii isolates were in 6 of 55 STs defined for C. sakazakii (4). However, there was an uneven distribution according to clinical records: all 5 cerebrospinal fluid (CSF) isolates were either ST4 or within the ST4 complex (clonal group where strains are identical in 4 or more loci). This group included strains from cases during December in Illinois (specimen 1577) and in Lebanon, Missouri (specimen 1579).
Specimen 1577 (ST110), isolated from CSF, is a triple-loci variant of ST4, distinguished by 5/3036 nt: atpD (1/390nt), gltB (2/507nt), and gyrB (2/402nt). Specimen 1578 (ST111), isolated from the PIF reconstitution water associated with the case reported in Illinois, is distinguishable from ST4 in 4/7 loci: fusA (5/438nt), glnS (1/363), infB (4/441), and ppsA (19/495). The 2 Illinois strains, 1577 and 1578 (ST110 and ST111), differed from each other at all loci, in total, 35/3,036 nt difference.
Such sequence-based relationship analysis of isolates is not possible by using pulsed-field gel electrophoresis (PFGE). PFGE and MLST analyze the bacterial DNA content differently, and there are no XbaI sites (the endonuclease most commonly used with PFGE of Enterobacteriaceae) within the 7 MLST loci. C. sakazakii ST4 strains were also found in feces (specimen 1567), opened PIF (specimen 1571), and tracheal samples (specimen 1576) (Figure). In addition, 2 single-loci ST4 variants were found; CSF specimen 1565 differed from the ST4 profile in the fusA loci by 6/438 nt, and specimen1572 from an opened tin of PIF differed in the fusA loci by 5/438 nt. These 2 strains differ from each other minimally, by 1 nt of 3,036 (concatenated length) in the fusA loci position 378 (A:T).
Several non-ST4 C. sakazakii strains were received by CDC in 2011. C. sakazakii ST8 was isolated from an opened powdered infant formula tin (specimen 1573) and 2 associated fecal samples from an infant who had diarrhea (specimens 1574 and 1575). One blood isolate (specimen 1569) was C. malonaticus ST112, found in an infant <1 month of age with meningitis who did not survive the infection. This finding is highly noteworthy because it has been proposed that C. malonaticus predominates in adult infections (5), and no fatal meningitis cases have been attributed to this species.
This MLST analysis of 15 strains received by the CDC in 2011 reinforces the conclusion that CSF isolates are not evenly spread across the 7 Cronobacter species and are instead predominantly in the C. sakazakii ST4 clonal complex. Such infections in neonates are of high concern because of the risk for associated severe brain damage. As previously stated, whether this association is caused by greater neonatal exposure as a result of environmental factors or particular virulence capabilities remains uncertain (5).
Acknowledgments
We thank Judith Noble-Wang and Matthew Arduino for their kind provision of strains.
Funding for this research was provided by Nottingham Trent University and Saudi Cultural Bureau.
References
- Baldwin A, Loughlin M, Caubilla-Barron J, Kucerova E, Manning G, Dowson C, Multilocus sequence typing of Cronobacter sakazakii and Cronobacter malonaticus reveals stable clonal structures with clinical significance which do not correlate with biotypes. BMC Microbiol. 2009;9:223. DOIPubMedGoogle Scholar
- Joseph S, Desai P, Ji Y, Cummings CA, Shih R, Degoricij L, Comparative analysis of genome sequences covering the seven Cronobacter species. PLoS ONE. 2012;7:e49455. DOIPubMedGoogle Scholar
- Joseph S, Cetinkaya E, Drahovska H, Levican A, Figueras M, Forsythe SJ. Cronobacter condimenti sp. nov., isolated from spiced meat and Cronobacter universalis sp. nov., a novel species designation for Cronobacter sp. genomospecies 1, recovered from a leg infection, water and food ingredients. Int J Syst Evol Microbiol. 2012;62:1277–83. DOIPubMedGoogle Scholar
- Joseph S, Sonbol H, Hariri S, Desai P, McClelland M, Forsythe SJ. Diversity of the Cronobacter genus as revealed by multi locus sequence typing. J Clin Microbiol. 2012;50:3031–9. DOIPubMedGoogle Scholar
- Joseph S, Forsythe SJ. Predominance of Cronobacter sakazakii ST4 in neonatal infections. Emerg Infect Dis. 2011;17:1713–5. DOIPubMedGoogle Scholar
- Centers for Disease Control and Prevention. CDC Update: investigation of Cronobacter infections among infants in the United States. January 13, 2012 [cited 2012 Aug 22]. http://www.cdc.gov/foodsafety/diseases/cronobacter/investigation.html
- Kucerova E, Joseph S, Forsythe S. The Cronobacter genus: ubiquity and diversity. Quality Assurance and Safety of Crops & Foods. 2011;3:104–22.
Figure
Table
Cite This ArticleRelated Links
Table of Contents – Volume 19, Number 1—January 2013
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Stephen Forsythe, School of Science and Technology, Nottingham Trent University, Clifton Lane, Nottingham NG11 8NS, UK
Top