Volume 20, Number 12—December 2014
Dispatch
Novel Porcine Epidemic Diarrhea Virus Variant with Large Genomic Deletion, South Korea
Figure
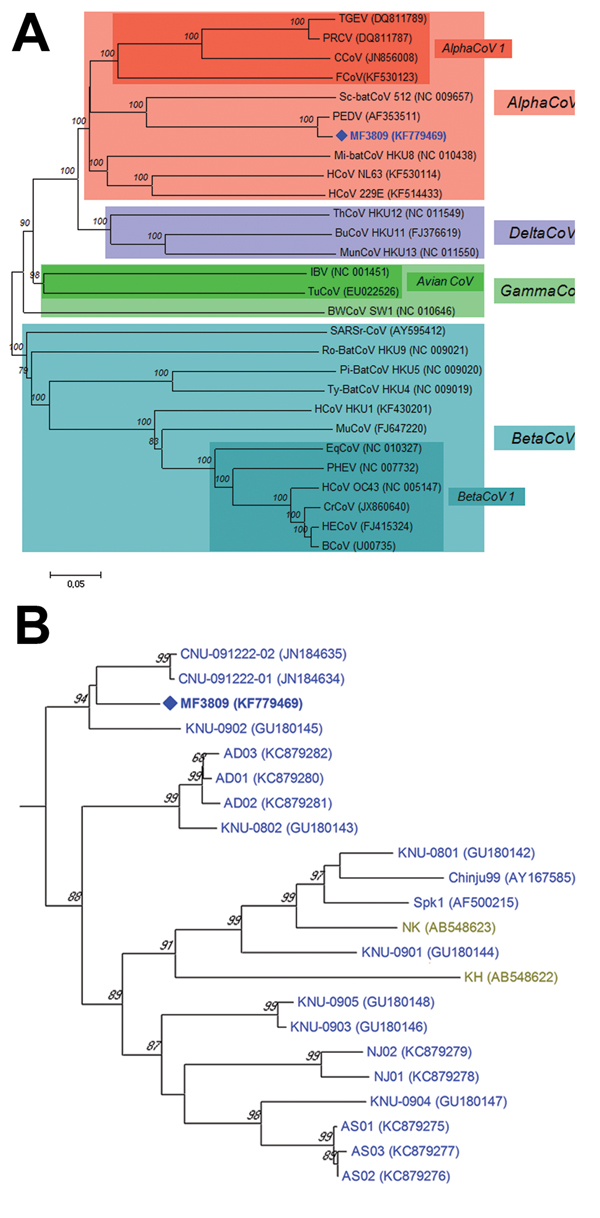
Figure. A) Relationships between the PEDV variant (MF3809/2008/South Korea) and other coronaviruses based on the full-length spike gene. PEDV, porcene epidemic diarrhea virus; TGEV, transmissible gastroenteritis virus; PRCV, porcine respiratory coronavirus; CCoV, canine coronavirus; FCoV, feline coronavirus; HCoV, human coronavirus; Mi-batCoV, Miniopterus bat coronavirus; Sc-batCoV, Scotophilus bat coronavirus; HECoV, human enteric coronavirus; BCoV, bovine coronavirus; PHEV, porcine hemagglutinating encephalomyelitis virus; EqCoV, equine coronavirus; CrCoV, canine respiratory coronavirus; MuCoV, murine coronavirus; Pi-BatCoV, Pipistrellus bat coronavirus; Ro-BatCoV, Rousettus bat coronavirus; SARSr-CoV, severe acute respiratory syndrome-related coronavirus; Ty-BatCoV, Tylonycteris bat coronavirus; IBV, infectious bronchitis virus; TuCoV, turkey coronavirus; BWCoV, Beluga whale coronavirus; BuCoV, bulbul coronavirus; ThCoV, thrush coronavirus; MunCoV, munia coronavirus. B) Phylogenetic tree of the entire spike genes of the PEDV variant and all known PEDV strains available in GenBank. The phylogenetic tree was constructed using the neighbor-joining clustering method in MEGA version 5.22 with a pairwise distance (5). Bootstrap values (based on 1,000 replicates) for each node are given if >60%. Scale bar indicates nucleotide substitutions per site. PEDV strains isolated from various countries are marked with colors as follows: Europe (black), China (red), Japan (olive green), USA (bright magenta) and South Korea (blue). PEDV, porcine epidemic diarrhea virus.
References
- Song D, Park B. Porcine epidemic diarrhoea virus: a comprehensive review of molecular epidemiology, diagnosis, and vaccines. Virus Genes. 2012;44:167–75. DOIPubMedGoogle Scholar
- Saif LJ, Pensaert MB, Sestak K, Yeo SG, Jung K. Coronaviruses. In: Zimmerman JJ, Karriker LA, Ramirez A, Schwartz KJ, Stevenson GW, editors. Disease of swine. Hoboken (NJ): Wiley-Blackwell; 2012. p. 501–24.
- Stevenson GW, Hoang H, Schwartz KJ, Burrough ER, Sun D, Madson D, Emergence of porcine epidemic diarrhea virus in the United States: clinical signs, lesions, and viral genomic sequences. J Vet Diagn Invest. 2013;25:649–54. DOIPubMedGoogle Scholar
- Park SJ, Song DS, Ha GW, Park BK. Cloning and further sequence analysis of the spike gene of attenuated porcine epidemic diarrhea virus DR13. Virus Genes. 2007;35:55–64. DOIPubMedGoogle Scholar
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–9. DOIPubMedGoogle Scholar
- Tian Y, Yu Z, Cheng K, Liu Y, Huang J, Xin Y, Molecular characterization and phylogenetic analysis of new variants of the porcine epidemic diarrhea virus in Gansu, China in 2012. Viruses. 2013;5:1991–2004. DOIPubMedGoogle Scholar
- Park SJ, Kim HK, Song DS, Moon HJ, Park BK. Molecular characterization and phylogenetic analysis of porcine epidemic diarrhea virus (PEDV) field isolates in Korea. Arch Virol. 2011;156:577–85. DOIPubMedGoogle Scholar
- Kim SH, Kim IJ, Pyo HM, Tark DS, Song JY, Hyun BH. Multiplex real-time RT-PCR for the simultaneous detection and quantification of transmissible gastroenteritis virus and porcine epidemic diarrhea virus. J Virol Methods. 2007;146:172–7. DOIPubMedGoogle Scholar
- Masters PS. The molecular biology of coronaviruses. Adv Virus Res. 2006;66:193–292. DOIPubMedGoogle Scholar
- Laude H, Van Reeth K, Pensaert M. Porcine respiratory coronavirus: molecular features and virus–host interactions. Vet Res. 1993;24:125–50 .PubMedGoogle Scholar
- Bernard S, Laude H. Site-specific alteration of transmissible gastroenteritis virus spike protein results in markedly reduced pathogenicity. J Gen Virol. 1995;76:2235–41. DOIPubMedGoogle Scholar
- Gallagher TM, Buchmeier MJ. Coronavirus spike proteins in viral entry and pathogenesis. Virology. 2001;279:371–4. DOIPubMedGoogle Scholar
- Callebaut P, Correa I, Pensaert M, Jimenez G, Enjuanes L. Antigenic differentiation between transmissible gastroenteritis virus of swine and a related porcine respiratory coronavirus. J Gen Virol. 1988;69:1725–30. DOIPubMedGoogle Scholar
- Reguera J, Ordono D, Santiago C, Enjuanes L, Casasnovas JM. Antigenic modules in the N-terminal S1 region of the transmissible gastroenteritis virus spike protein. J Gen Virol. 2011;92:1117–26. DOIPubMedGoogle Scholar
- Sánchez CM, Gebauer F, Suñé C, Mendez A, Dopazo J, Enjuanes L. Genetic evolution and tropism of transmissible gastroenteritis coronaviruses. Virology. 1992;190:92–105. DOIPubMedGoogle Scholar
1These authors were co-principal investigators for this study.