Volume 20, Number 6—June 2014
Research
Bats as Reservoir Hosts of Human Bacterial Pathogen, Bartonella mayotimonensis
Abstract
A plethora of pathogenic viruses colonize bats. However, bat bacterial flora and its zoonotic threat remain ill defined. In a study initially conducted as a quantitative metagenomic analysis of the fecal bacterial flora of the Daubenton’s bat in Finland, we unexpectedly detected DNA of several hemotrophic and ectoparasite-transmitted bacterial genera, including Bartonella. Bartonella spp. also were either detected or isolated from the peripheral blood of Daubenton's, northern, and whiskered bats and were detected in the ectoparasites of Daubenton's, northern, and Brandt's bats. The blood isolates belong to the Candidatus-status species B. mayotimonensis, a recently identified etiologic agent of endocarditis in humans, and a new Bartonella species (B. naantaliensis sp. nov.). Phylogenetic analysis of bat-colonizing Bartonella spp. throughout the world demonstrates a distinct B. mayotimonensis cluster in the Northern Hemisphere. The findings of this field study highlight bats as potent reservoirs of human bacterial pathogens.
The 1,100 species of bats (1) constitute ≈20% of known mammalian species and are outnumbered only by animals in the order Rodentia. Bats play a vital role in natural ecosystems in arthropod suppression, seed dispersal, and pollination. Modern-day economies also benefit from these voracious predators of crop and forest pests (2). However, bats have been implicated as reservoir hosts for viral human pathogens, such as paramyxoviruses (3) and rabies virus and related lyssaviruses (4). Compelling evidence also indicates that bats carry asymptomatically some of the most deadly viruses, including Marburg (5) and Ebola (6) viruses. Whether bats carry clinically significant bacterial pathogens is unknown.
The development of next-generation sequencing techniques has revolutionized biological science. It is now possible—and cost-friendly—to gain access to massive amounts of qualitative and quantitative sequencing data in a short time without a priori knowledge of the sequence (7). Most bacteria do not grow on laboratory media, and next-generation sequencing technologies have proven useful for studying bacterial species diversity and dynamics, even in complex systems like the gut (8). Our initial objective in 2010 and 2011 was to conduct a quantitative metagenomic analysis of the fecal bacterial flora of the Daubenton’s bat (Myotis daubentonii) in Finland. Unexpectedly, we found that the fecal material contained DNA of several hemothrophic and ectoparasite-transmitted bacterial genera, such as Bartonella. This DNA may originate either from bleeding into the intestine or from the insect prey of the bats that includes the abundant bloodfeeding bat ectoparasites. Therefore, the study further focused on detecting and isolating Bartonella spp. from peripheral blood and ectoparasites of several bat species in Finland in 2012.
Bartonella spp. nucleotide sequences have been deposited in GenBank under accession nos. KF003115–KF003145. The metagenomic reads are stored at the National Center for Biotechnology Information Sequence Read Archive under BioProject SRP023235 (accession nos. experiment: SRX286839, run: SRR868695). We have described the detailed protocols, including bat sampling for peripheral blood, fecal droppings, and ectoparasites; metagenomic analysis of fecal DNA; isolation of Bartonella from peripheral blood; extraction of DNA from bat blood, ectoparasites, and Bartonella isolates; Bartonella and ectoparasite PCR analyses; transmission electron microscopy; and nucleotide sequence and phylogenetic analyses in the Technical Appendix.
Quantitative Metagenomic Analysis of DNA from Bat Feces
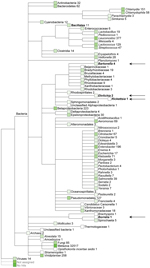
We obtained ≈200,000 high-quality sequences (average length 167 bp) from DNA sequencing of fecal material from a Daubenton’s bat (Technical Appendix Figure 1). Sequences (>50 bp) were assigned on the basis of best E-value BLASTN scores (http://blast.ncbi.nlm.nih.gov/blast.cgi) in GenBank. The most abundant non-metazoan sequence matches were with bacteria. The genera Leuconostoc, Enterobacter, Lactococcus, and Chlamydia dominated (Figure 1). Surprisingly, the fecal material also contained DNA of the ectoparasite-transmitted genera, such as the hemotrophic bartonellae (9). It was thought that this DNA originated either from bleeding into the intestine or from the insect prey of the bats that includes the abundant bloodfeeding bat ectoparasites. PCR verified the presence of Bartonella DNA in the bat fecal material. The transfer messenger RNA gene (ssrA) (10) could be amplified and was sequenced from the fecal material of 1 Daubenton’s bat, 1 northern bat (Eptesicus nilssonii), and 1 Brandt’s bat (Myotis brandtii) (no. 2771, no. 2788, and no. 2786, respectively; Technical Appendix Table 1). The obtained 218-bp ssrA sequences were 100% identical. The closest matches in GenBank, with a similarity score of 94.8% (183/193 bp), were B. tamiae Th339 (GenBank accession no. JN029780) and Th307 strains (GenBank accession no. JN029778) isolated from 2 humans in Thailand (11).
Candidatus Status Species B. mayotimonensis and Novel Bartonella Species
Bats belonging to the 4 most prevalent bat species in Finland were captured in August and September 2012 at 3 locations in southwestern Finland (Technical Appendix Table 1). Culturing of peripheral blood samples of 5 Daubenton’s bats and 1 northern bat yielded distinct colonies. The isolates were identified as Bartonella spp. by sequencing a PCR-amplified 485-bp fragment containing the hypervariable regions V6–V8 of the 16S rRNA gene. Overall health of the bats as analyzed by body condition indexing was not affected by the Bartonella infection (Technical Appendix Table 1).
The 16S rRNA gene sequences are highly conserved within the genus Bartonella and thus not robust in differentiating species (12). Therefore, we sequenced PCR-amplified fragments of the RNA polymerase β-subunit gene (rpoB), citrate synthase gene (gltA), filamenting temperature-sensitive mutant Z gene (ftsZ), VirB type IV secretion system VirB4 component gene (virB4), hypervariable region 2 of the 16S-23S rRNA intergenic spacer region (ISR), and ssrA (Technical Appendix Table 2). Sequencing of rpoB was first conducted on all 28 clonal isolates. Three distinct rpoB alleles were identified (Technical Appendix Table 1). The multilocus sequence analysis (MLSA) was completed on 1 rpoB-1 allele isolate (clone 3, bat no. 1157, referred to hereafter as 1157/3), 1 rpoB-2 allele isolate (clone 1, bat no. 2574, referred to hereafter as 2574/1), and 1 rpoB-3 allele isolate (clone 1, bat no. 1160, referred to hereafter as 1160/1). Thin-section transmission electron micrographs of these isolates are shown in the Technical Appendix Figure 2. No major pili or fimbriae-like structures were detected on the surface of the rod-shaped bacteria.
Results of BLASTN homology searches performed in January 2013 are shown in Technical Appendix Table 3. ISR is a robust species discriminatory marker within the genus Bartonella (13,14). ISR of the strain 2574/1 did not have any hits, whereas ISR of strains 1157/3 and 1160/1 had on1y 1 hit in GenBank Candidatus B. mayotimonensis (15), with high sequence similarity scores. Sequence analyses of the other MLSA markers (Technical Appendix Table 3) further indicate that isolates 1157/3 and 1160/1 belong to the Candidatus-status species B. mayotimonensis and that strain 2574/1 belongs to a new Bartonella species. Indeed, the lowest pairwise genetic distance values with the concatenated rpoB, gltA, 16S rRNA, and ftsZ sequence fragments of the bat strains 1157/3 and 1160/1 in the genus Bartonella were 0.040 and 0.038, respectively, with Candidatus B. mayotimonensis (Technical Appendix Table 4). Because the distance value 0.05 is the recommended cutoff value for species delineation (16), the bat isolates 1157/3 and 1160/1 classify as strains of the Candidatus-status species B. mayotimonensis. The bat strain 2574/1 belongs to a new Bartonella species because the lowest genetic distance value in the genus Bartonella was 0.070 with B. washoensis, above the 0.05 cutoff value (16).
Figure 2 shows the phylogenetic position of the bat Bartonella isolates based on comparisons of concatenated sequences of rpoB, gltA, 16S rRNA and ftsZ, available for Candidatus B. mayotimonensis (15) and all type strains of the Bartonella species (Technical Appendix Table 5). The neighbor-joining and maximum-likelihood trees demonstrate that bat isolates 1157/3 and 1160/1 cluster with Candidatus B. mayotimonensis with high bootstrap values in a distinct phylogenetic position. The new Bartonella species (strain 2754/1) clearly diverges from the other bat isolates.
Bat Ectoparasite Flies and Fleas as Vectors for Transmitting Bartonella
We sequenced a PCR-amplified fragment of the mitochondrial cytochrome c oxidase subunit I (17) and also used visual inspection to identify the ectoparasites of 18 bats (o Technical Appendix Table 1). Ectoparasite DNA preparations of 2 fleas and 10 flies were analyzed with a PCR protocol targeting the Bartonella rpoB. The blood isolate rpoB alleles 1 and 2 were detected in samples from 1 flea and 2 flies, respectively (Technical Appendix Table 1). In addition, 2 novel rpoB alleles were detected. The rpoB-5 allele detected in a fly sample is distantly related to the currently known Bartonella rpoB sequences. The highest BLASTN sequence identity score with the rpoB-4 allele detected in a flea sample, and from 1 blood DNA preparation of a culture-negative whiskered bat (no. 1156, Technical Appendix Table 1), was 97.8% (397/406 bp) with the corresponding fragment (FJ376736) of Candidatus B. mayotimonensis. This is a higher value than with the rpoB-1 and rpoB-2 alleles. Moreover, a partial 338-bp gltA fragment could be amplified from the rpoB-4–positive flea sample. The highest BLASTN sequence identity score with Candidatus B. mayotimonensis was 93.2% (315/338 bp), which is higher than with the isolates 1157/3 (92.0%, 311/338 bp) and 1160/1 (92.3%, 312/338 bp). The data further support the conclusion that bats are reservoir hosts of B. mayotimonensis and indicate that the bat flies and fleas transmit Bartonella spp. to new hosts.
Phylogenetic Analysis of Bartonella spp. that Colonize Bats Worldwide
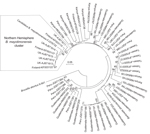
A maximum composite likelihood–based neighbor-joining tree (Figure 3) was constructed on the basis of 253-bp gltA sequences obtained from Bartonella that infect bats in the United Kingdom (18), Kenya (19), Guatemala (20), Taiwan (21), and Peru (22). The 5 Bartonella-like bacteria detected in minced heart tissues in the United Kingdom (18), the B. mayotimonensis isolates from Finland, and the strain detected in 1 bat flea in Finland clustered in a distinct phylogenetic position away from the bat isolates and strains of the Southern Hemisphere. The new Bartonella species (strain 2574/1) does not belong to the Northern Hemisphere B. mayotimonensis cluster. Remarkably, the 253-bp gltA fragment of 1 of the Bartonella-like bacteria detected in minced heart tissue of a common noctule (Nyctalus noctule) (Cornwall-M451, AJ871615) yielded a 95.3% (241/253 bp) sequence identity score, compared with the corresponding fragment (FJ376732) of Candidatus B. mayotimonensis. This value is significantly higher than those obtained with corresponding gltA fragments of Finland bat isolates 1157/3 (92.9%, 235/253) or 1160/1 (94.1%, 238/253). The UK gltA data further support the conclusion that bats are reservoir hosts of B. mayotimonensis. Most importantly, bats appear to be reservoir hosts of B. mayotimonensis only in the Northern Hemisphere.
Bartonella naantaliensis (naan.tali´en.sis. N.L. fem. adj. n. naantaliensis of or belonging to Naantali) is the name proposed to highlight the municipality where the bat was trapped from which the type strain was isolated. The type strain is 2574/1. Its partial 16S rRNA gene nucleotide sequence is deposited in GenBank (accession no. KF003116).
Bartonella spp. are facultative intracellular bacteria that typically cause long-lasting hemotrophic bacteremia in their mammalian reservoir hosts, such as rodents (9). The relapsing bacteremia can last weeks, months, or even years, thereby favoring transmission by bloodfeeding arthropods. In recent years, increasing numbers of Bartonella spp. have been implicated as zoonotic human pathogens. A frequent symptom is endocarditis, usually suspected in cases in which conventional culture-based diagnostics fail. The most prevalent endocarditis-causing species are B. quintana (23,24) and B. henselae (25), but B. elizabethae (26), B. alsatica (27), B. koehlerae (28), B. vinsonii subsp. berkhoffii (29), and B. vinsonii subsp. arupensis (30) also have been detected or isolated. Recently, a new type of Bartonella was detected in a resected aortic valve tissue of a human endocarditis patient (15). A species name, Candidatus B. mayotimonensis was proposed because a pure microbiological culture was not obtained. The reservoir host in nature also remained elusive. As part of a study designed to characterize the microbiome of bats, bacteria that belong to the Candidatus-status species B. mayotimonensis were either detected or isolated from peripheral blood samples and the ectoparasites of bats. In addition, a new Bartonella species (strain 2574/1) was isolated from the blood and detected from the ectoparasites.
The ad hoc committee to reevaluate the species definition in bacteriology has proposed that descriptions of novel species could be based solely on gene sequence analyses (31). In the current study, 6 genes, including the robust Bartonella spp. discriminatory marker, the ISR, were used (13,14). It is remarkable that ISR of the 2574/1 isolate did not have any hits, whereas ISRs of 1157/3 and 1160/1 isolates had only 1 hit in GenBank, Candidatus B. mayotimonensis. If gltA shares <96.0% and rpoB <95.4% nt sequence similarity with those of the validated species, the newly encountered Bartonella strain can be considered a new species (32). According to these criteria, which were proposed in 2003 when half of the currently known species were known, the bat isolate 2574/1 is a new Bartonella species. The bat isolates 1157/3 and 1160/1 belong to the Candidatus-status species B. mayotimonensis on the basis of the rpoB sequences but would belong to a new Bartonella species on the basis of the gltA sequences. Because the species classification gave contradictory results, sequence analyses of other MLSA markers and phylogenetic analyses were performed. In addition, we used 4 concatenated MLSA markers to determine pairwise genetic distance values to the known members of the genus. The bat isolate 1157/3 and 1160/1 ftsZ sequences had a significantly higher sequence similarity with ftsZ of Candidatus B. mayotimonesis than with any other type strain sequence. The neighbor-joining and maximum-likelihood phylogenetic trees with the concatenated rpoB, gltA, 16S rRNA and ftsZ sequences both demonstrated that the bat isolates 1157/3 and 1160/1 cluster with Candidatus B. mayotimonensis with high bootstrap values in a distinct phylogenetic position. Moreover, the genetic distance values demonstrate that the bat isolates 1157/3 and 1160/1 classify as strains of the Candidatus-status species B. mayotimonensis. We propose that the bat isolate 1160/1 is the type strain of B. mayotimonensis.
Findings of the study raised an interesting question: how could Bartonella spp., or any other hemotrophic bacterium, be transmitted from the bat into the human host? Daubenton’s bats prefer to roost in abandoned woodpecker cavities and bird boxes, whereas the other bat species are often found in the attics of houses in close proximity to humans. Given that Bartonella spp. are hemotrophic, transmission through bat bite and saliva is not considered likely. Moreover, at Turku University Central Hospital, which is responsible for a population base of 500,000, only 2 or 3 patients per year are admitted with a bat bite (J. Oksi, pers. comm.). These numbers probably reflect the frequency of bat bites in most countries of the Northern Hemisphere. We propose that fecal droppings of blood-fed bat ectoparasites might transmit Bartonella spp. into the human host, assisted by superficial scratching or tissue trauma of the skin. The presence of viable bacteria in feces of body lice (Pediculus humanus) that have been feeding on B. quintana–infected rabbits is well documented (33,34). Similar observations have been reported for the feces of experimentally infected cat fleas (Ctenocephalides felis) (35,36). Most importantly, intradermal injection of feces from fleas that had fed on a B. henselae–infected cat led to bacteremia in a pathogen-free cat (37). Ectoparasite bite–mediated transmission is also possible, but the bat bugs (Cimex spp.) known to also feed on humans were not analyzed in the current study.
The reported metagenomic analysis of bat fecal material indicates that bats are reservoir hosts for several pathogenic bacterial genera. No comprehensive study has been published on the bacterial flora of bats in light of its zoonotic threat to humans. The major research focus has been on viruses, and several deadly viruses have been detected or isolated (3–6). One of the main conclusions from these studies is that bats tolerate their deadly companions relatively well, a feature that has been discussed in the context of long evolutionary history of bats (38). Bats are also highly mobile and long-lived, ideal as pathogen reservoirs. Metagenomics-driven approaches should be continued to assess the pathogenic potential of bacteria that colonize bats.
Dr Veikkolainen is a postdoctoral research fellow of the Pulliainen laboratory. His research interests include the dynamics of bat microbiome and bat immunology, with special emphasis on the recognition and control of bacterial colonization.
Mr Vesterinen is a PhD student at the Department of Biology, University of Turku. His research interests include food web dynamics and biological interactions, and he has established a laboratory dedicated for fecal analysis in the Laboratory of Genetics, University of Turku.
Acknowledgments
We gratefully thank Olaf Thalmann for help in the laboratory.
This work was supported by grant 8149 and grant 9222 from the Turku University Foundation to A.T.P. and by personal grant from Emil Aaltonen Foundation and grant 8621 from Turku University Foundation to E.J.V. We acknowledge CSC–IT Center for Science Ltd. for the allocation of computational resources.
References
- Schipper J, Chanson JS, Chiozza F, Cox NA, Hoffmann M, Katariya V, The status of the world's land and marine mammals: diversity, threat, and knowledge. Science. 2008;322:225–30. DOIPubMedGoogle Scholar
- Boyles JG, Cryan PM, McCracken GF, Kunz TH. Conservation. Economic importance of bats in agriculture. Science. 2011;332:41–2. DOIPubMedGoogle Scholar
- Drexler JF, Corman VM, Müller MA, Maganga GD, Vallo P, Binger T, Bats host major mammalian paramyxoviruses. Nat Commun. 2012;3:796. Erratum in: Nat Commun. 2014;5:3032.
- Rupprecht CE, Turmelle A, Kuzmin IV. A perspective on lyssavirus emergence and perpetuation. Curr Opin Virol. 2011;1:662–70.
- Towner JS, Amman BR, Sealy TK, Carroll SA, Comer JA, Kemp A, Isolation of genetically diverse Marburg viruses from Egyptian fruit bats. PLoS Pathog. 2009;5:e1000536. DOIPubMedGoogle Scholar
- Leroy EM, Kumulungui B, Pourrut X, Rouquet P, Hassanin A, Yaba P, Fruit bats as reservoirs of Ebola virus. Nature. 2005;438:575–6. DOIPubMedGoogle Scholar
- Shokralla S, Spall JL, Gibson JF, Hajibabaei M. Next-generation sequencing technologies for environmental DNA research. Mol Ecol. 2012;21:1794–805. DOIPubMedGoogle Scholar
- Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–14. DOIPubMedGoogle Scholar
- Pulliainen AT, Dehio C. Persistence of Bartonella spp. stealth pathogens: from subclinical infections to vasoproliferative tumor formation. FEMS Microbiol Rev. 2012;36:563–99. DOIPubMedGoogle Scholar
- Diaz MH, Bai Y, Malania L, Winchell JM, Kosoy MY. Development of a novel genus-specific real-time PCR assay for detection and differentiation of Bartonella species and genotypes. J Clin Microbiol. 2012;50:1645–9. DOIPubMedGoogle Scholar
- Kosoy M, Morway C, Sheff KW, Bai Y, Colborn J, Chalcraft L, Bartonella tamiae sp. nov., a newly recognized pathogen isolated from three human patients from Thailand. J Clin Microbiol. 2008;46:772–5. DOIPubMedGoogle Scholar
- Kosoy M, Hayman DT, Chan KS. Bartonella bacteria in nature: where does population variability end and a species start? Infect Genet Evol. 2012;12:894–904. DOIPubMedGoogle Scholar
- García-Esteban C, Gil H, Rodríguez-Vargas M, Gerrikagoitia X, Barandika J, Escudero R, Molecular method for Bartonella species identification in clinical and environmental samples. J Clin Microbiol. 2008;46:776–9 . DOIPubMedGoogle Scholar
- Houpikian P, Raoult D. 16S/23S rRNA intergenic spacer regions for phylogenetic analysis, identification, and subtyping of Bartonella species. J Clin Microbiol. 2001;39:2768–78 . DOIPubMedGoogle Scholar
- Lin EY, Tsigrelis C, Baddour LM, Lepidi H, Rolain JM, Patel R, Candidatus Bartonella mayotimonensis and endocarditis. Emerg Infect Dis. 2010;16:500–3. DOIPubMedGoogle Scholar
- Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 2007;57:81–91. DOIPubMedGoogle Scholar
- Zeale MR, Butlin RK, Barker GL, Lees DC, Jones G. Taxon-specific PCR for DNA barcoding arthropod prey in bat faeces. Mol Ecol Resour. 2011;11:236–44.
- Concannon R, Wynn-Owen K, Simpson VR, Birtles RJ. Molecular characterization of haemoparasites infecting bats (Microchiroptera) in Cornwall, UK. Parasitology. 2005;131:489–96. DOIPubMedGoogle Scholar
- Kosoy M, Bai Y, Lynch T, Kuzmin IV, Niezgoda M, Franka R, Bartonella spp. in bats, Kenya. Emerg Infect Dis. 2010;16:1875–81. DOIPubMedGoogle Scholar
- Bai Y, Kosoy M, Recuenco S, Alvarez D, Moran D, Turmelle A, Bartonella spp. in bats, Guatemala. Emerg Infect Dis. 2011;17:1269–72. DOIPubMedGoogle Scholar
- Lin JW, Hsu YM, Chomel BB, Lin LK, Pei JC, Wu SH, Identification of novel Bartonella spp. in bats and evidence of Asian gray shrew as a new potential reservoir of Bartonella. Vet Microbiol. 2012;156:119–26. DOIPubMedGoogle Scholar
- Bai Y, Recuenco S, Gilbert AT, Osikowicz LM, Gómez J, Rupprecht C, Prevalence and diversity of Bartonella spp. in bats in Peru. Am J Trop Med Hyg. 2012;87:518–23. DOIPubMedGoogle Scholar
- Drancourt M, Mainardi JL, Brouqui P, Vandenesch F, Carta A, Lehnert F, Bartonella (Rochalimaea) quintana endocarditis in three homeless men. N Engl J Med. 1995;332:419–23. DOIPubMedGoogle Scholar
- Chaloner GL, Harrison TG, Birtles RJ. Bartonella species as a cause of infective endocarditis in the UK. Epidemiol Infect. 2013;141:841–6 . DOIPubMedGoogle Scholar
- Holmes AH, Greenough TC, Balady GJ, Regnery RL, Anderson BE, O'Keane JC, Bartonella henselae endocarditis in an immunocompetent adult. Clin Infect Dis. 1995;21:1004–7. DOIPubMedGoogle Scholar
- Daly JS, Worthington MG, Brenner DJ, Moss CW, Hollis DG, Weyant RS, Rochalimaea elizabethae sp. nov. isolated from a patient with endocarditis. J Clin Microbiol. 1993;31:872–81 .PubMedGoogle Scholar
- Raoult D, Roblot F, Rolain JM, Besnier JM, Loulergue J, Bastides F, First isolation of Bartonella alsatica from a valve of a patient with endocarditis. J Clin Microbiol. 2006;44:278–9. DOIPubMedGoogle Scholar
- Avidor B, Graidy M, Efrat G, Leibowitz C, Shapira G, Schattner A, Bartonella koehlerae, a new cat-associated agent of culture-negative human endocarditis. J Clin Microbiol. 2004;42:3462–8. DOIPubMedGoogle Scholar
- Roux V, Eykyn SJ, Wyllie S, Raoult D. Bartonella vinsonii subsp. berkhoffii as an agent of afebrile blood culture-negative endocarditis in a human. J Clin Microbiol. 2000;38:1698–700 .PubMedGoogle Scholar
- Fenollar F, Sire S, Wilhelm N, Raoult D. Bartonella vinsonii subsp. arupensis as an agent of blood culture-negative endocarditis in a human. Erratum in: J Clin Microbiol. 2005;43:4923. J Clin Microbiol. 2005;43:945–7.
- Stackebrandt E, Frederiksen W, Garrity GM, Grimont PA, Kämpfer P, Maiden MC, Report of the ad hoc committee for the re-evaluation of the species definition in bacteriology. Int J Syst Evol Microbiol. 2002;52:1043–7. DOIPubMedGoogle Scholar
- La Scola B, Zeaiter Z, Khamis A, Raoult D. Gene-sequence-based criteria for species definition in bacteriology: the Bartonella paradigm. Trends Microbiol. 2003;11:318–21. DOIPubMedGoogle Scholar
- Seki N, Kasai S, Saito N, Komagata O, Mihara M, Sasaki T, Quantitative analysis of proliferation and excretion of Bartonella quintana in body lice, Pediculus humanus L. Am J Trop Med Hyg. 2007;77:562–6 .PubMedGoogle Scholar
- Fournier PE, Minnick MF, Lepidi H, Salvo E, Raoult D. Experimental model of human body louse infection using green fluorescent protein-expressing Bartonella quintana. Infect Immun. 2001;69:1876–9. DOIPubMedGoogle Scholar
- Higgins JA, Radulovic S, Jaworski DC, Azad AF. Acquisition of the cat scratch disease agent Bartonella henselae by cat fleas (Siphonaptera:Pulicidae). J Med Entomol. 1996;33:490–5 .PubMedGoogle Scholar
- Finkelstein JL, Brown TP, O'Reilly KL, Wedincamp J, Foil LD. Studies on the growth of Bartonella henselae in the cat flea (Siphonaptera: Pulicidae). J Med Entomol. 2002;39:915–9. DOIPubMedGoogle Scholar
- Foil L, Andress E, Freeland RL, Roy AF, Rutledge R, Triche PC, Experimental infection of domestic cats with Bartonella henselae by inoculation of Ctenocephalides felis (Siphonaptera: Pulicidae) feces. J Med Entomol. 1998;35:625–8 .PubMedGoogle Scholar
- Zhang G, Cowled C, Shi Z, Huang Z, Bishop-Lilly KA, Fang X, Comparative analysis of bat genomes provides insight into the evolution of flight and immunity. Science. 2013;339:456–60. DOIPubMedGoogle Scholar
Figures
Cite This Article1These authors contributed equally to this article.
Table of Contents – Volume 20, Number 6—June 2014
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Arto T. Pulliainen, Division of General Microbiology, Department of Biosciences, University of Helsinki, P.O. Box 56 (Viikinkaari 9), Biocenter 1, FI 00790, Helsinki, Finland,
Top