Volume 21, Number 2—February 2015
Dispatch
Comparative Analysis of African Swine Fever Virus Genotypes and Serogroups
Abstract
African swine fever virus (ASFV) causes highly lethal hemorrhagic disease among pigs, and ASFV’s extreme antigenic diversity hinders vaccine development. We show that p72 ASFV phylogenetic analysis does not accurately define ASFV hemadsorption inhibition assay serogroups. Thus, conventional ASFV genotyping cannot discriminate between viruses of different virulence or predict efficacy of a specific ASFV vaccine.
African swine fever (ASF) is a highly contagious hemorrhagic disease that causes high rates of death among domestic pigs. The disease is caused by ASF virus (ASFV), and the extreme antigenic diversity of the virus is one of the main obstacles to developing a safe and efficacious vaccine against ASF. Nevertheless, substantial progress has been made in understanding the pathogenesis of the disease and virus–host interactions (1,2). The ability to induce a protective immune response against ASFV has been demonstrated in numerous studies. Pigs that recover from ASF have long-term immunity to subsequent challenge with moderately virulent ASFV and related virulent viruses, but they rarely gain immunity to heterologous viruses (3–6). Because of these cross-protective responses, the antigenic diversity among naturally occurring ASFV isolates is of interest for ASFV vaccine development (7).
Researchers at the National Research Institute for Veterinary Virology and Microbiology (VNIIVViM) in Рokrov, Russia, have developed a classification of ASFV isolates based on a hemadsorption inhibition assay (HAI) with ASFV reference immune antisera. The results of a long-term study from VNIIVViM were used to serologically classify ASFV strains, isolates, and attenuated variants. Eight serogroups have been identified (serogroups 1–8), but more likely exist. In vaccine design and development, consideration should be given to the fact that viruses within a serogroup provide cross-protection from challenge with viruses of the same serogroup (8,9). VNIIVViM maintains a large and diverse collection of serologically grouped ASFV isolates that provides a unique resource for defining ASFV strain variability and establishing relationships of cross-protective immunity (10,11).
Current genetic typing of ASFV isolates is based on nucleotide sequencing of the p72 capsid protein gene (B646L) and/or amplification of full-length polymorphisms of various genomic regions (12,13). During ASF outbreaks, these genotyping approaches can be used to identify the origin of viruses and quickly differentiate closely related strains. However, the correlation between currently established ASFV genotypes and viral cross-protection is not precisely clear (6). Thus, we examined the relationship of the established genotype distribution to HAI serologic classification.
Serologic classification was based on HAI results for ASFV strains maintained at VNIIVViM. These include isolates from disease outbreaks in Africa, Europe, the Caribbean, and, more recently, from the Russian/Trans-Caucasian epizootic and attenuated variants. In brief, swine red bone marrow cell culture was used for ASFV isolate amplification, and swine anti-ASFV serum and erythrocytes were subsequently added to the culture. ASFV isolates for which the hemadsorption phenomenon was inhibited by serum belonging to the same group within serogroups 1–8 were clustered into a homologous serogroup. Only ASFV hemadsorbing strains could be analyzed by this method, and some hemadsorbing ASFV isolates could not be placed into existing serogroups because HAI was not observed with available reference serum (8,9).
ASFV isolates from the depository at VNIIVViM were also classified by using a standard ASFV genotyping protocol previously published by Bastos et al. (13). In this method, the variable part of the p72 (B646L) gene was amplified by conventional PCR, and the amplicons were directly sequenced by using a 3130xl Genetic Analyzer (Applied Biosystems, Foster City, CA, USA) according to manufacturer’s recommendations. Chromatograms were manually edited and assembled by using CAP3 (http://pbil.univ-lyon1.fr/cap3.php). The nucleotide sequences of the ASFV isolates were deposited into GenBank (accession nos. KJ526354–KJ5263471).
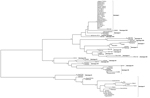
Sequences determined at VNIIVViM were aligned with other publically available ASFV sequences and analyzed by using minimum evolution; a rooted tree was constructed with MEGA 5.0 software (14) and edited with FigTree v1.4 (http://tree.bio.ed.ac.uk/) (Figure 1). The results of the ASFV genotyping are summarized in the Table.
Newly identified ASFV genotypes and known serogroups were mapped together so their geographic distribution in Africa and Europe (including the European part of the Russian Federation) could be visualized. The results (Figure 2) show that genotypic and serogroup diversity are greatest in a relatively limited area, mainly in southeastern Africa. In contrast, non-ASFV–endemic countries, where ASF outbreaks were caused by ASFV of a single genotype, exhibited low or no serogroup diversity.
Single genotype clades of ASFV were observed to contain viruses of multiple serogroups (Table). For example, ASFV isolates belonging to serogroups 1, 2, and 4 were specifically clustered within genotype I, and did not group with other genotypes. This indicates heterogeneity among ASFV strains previously isolated on the European continent.
We also found several serogroups of ASF viruses within genotype X. The ASFV isolates TSP80 (serogroup 5) and TS-7 (serogroup 6) were subsequently isolated from 1 field sample derived from a naturally infected pig in Tanzania. However both were genotype X viruses. Of note, 1 serogroup 2 isolate (Spenser) demonstrated a novel genotype within the p72 phylogenetic tree and relative to other serogroup 2 viruses (Figure 1), indicating that the p72 genotype, in addition to lacking serotype resolution, has potential to be incongruous relative to serogroup. Together, these data indicate that the antigenic heterogeneity of ASFV strains is not fully captured by using the standard genotyping approach.
The virus elements responsible for protective and cross-protective immune responses are not well known. Given the structural and genetic complexity of ASFV, it is likely that genes encoding different antigens will be more suited for virus typing. Substantial genetic variability can exist between strains and predominate in specific genomic regions, and it is these regions that may provide improved targets for genotyping. Our findings support that of a previous study that showed that geographic areas with ASFV of high genotypic and serotypic diversity are located in countries where multiple mechanisms of ASF transmission (mixed sylvatic and domestic cycle) are established (11).
HAI serology provides a measure of ASFV typing that, compared with p72 genotyping, better discriminates biologically pertinent phenotypes. Viruses belonging to one p72 genotype may be serotypically heterogeneous: strains that are closely related genetically, even from a single isolate, may have different phenotypes and form homologous serogroups. Our assessment of ASFV genotyping relating to HAI serotyping shows the serologic diversity within a p72 genotype. Our results highlight the potential for using serogroup classification to understand issues of homologous cross-protection among ASFV isolates and virus determinants that influence disease emergence.
The key finding from our study is that p72 ASFV phylogenetic analysis fails to accurately define ASFV HAI serogroups. Thus, conventional ASFV genotyping cannot discriminate between viruses of different virulence or predict the efficacy of a specific ASFV vaccine. This finding also suggests that for vaccination-based control of ASF, it may be more important to determine serogroups rather than genotypes of ASFV isolates. Nevertheless, more ASFV sampling is needed to further define genotypes and serogroups.
Dr Malogolovkin is a head of the molecular virology unit at the Institute of Veterinary Virology and Microbiology, Russian Academy of Agriculture Science. His research interests are the molecular evolution and genomics of African swine fever virus.
Acknowledgments
We thank Sodnom Tsybanov for helpful discussion; Edan Tulman for remarks and critical reading of the manuscript; and Alexandra Elsukova and Nadejda Knyazeva for technical assistance with the handling of ASFV isolates.
Financial support was provided by the Council of the President of the Russian Federation (grant no. MK-1843.2013.4) and the Russian Academy of Agriculture Science.
References
- Blome S, Gabriel C, Beer M. Pathogenesis of African swine fever in domestic pigs and European wild boar. Virus Res. 2013;173:122–30. DOIPubMedGoogle Scholar
- Correia S, Ventura S, Parkhouse RM. Identification and utility of innate immune system evasion mechanisms of ASFV. Virus Res. 2013;173:87–100. DOIPubMedGoogle Scholar
- Hamdy FM, Dardiri AH. Clinical and immunologic responses of pigs to African swine fever virus isolated from the Western Hemisphere. Am J Vet Res. 1984;45:711–4 .PubMedGoogle Scholar
- Lewis T, Zsak L, Burrage TG, Lu Z, Kutish GF, Neilan JG, An African swine fever virus ERV1-ALR homologue, 9GL, affects virion maturation and viral growth in macrophages and viral virulence in swine. J Virol. 2000;74:1275–85. DOIPubMedGoogle Scholar
- Moore DM, Zsak L, Neilan JG, Lu Z, Rock DL. The African swine fever virus thymidine kinase gene is required for efficient replication in swine macrophages and for virulence in swine. J Virol. 1998;72:10310–5 .PubMedGoogle Scholar
- King K, Chapman D, Argilaguet JM, Fishbourne E, Hutet E, Cariolet R, Protection of European domestic pigs from virulent African isolates of African swine fever virus by experimental immunisation. Vaccine. 2011;29:4593–600. DOIPubMedGoogle Scholar
- Dixon LK, Abrams CC, Chapman DD, Goatley LC, Netherton CL, Taylor G, Prospects for development of African swine fever virus vaccines. Dev Biol (Basel). 2013;135:147–57. DOIPubMedGoogle Scholar
- Vishnyakov I, Mitin N, Karpov G, Kurinnov V, Yashin A. Differentiation of African and classical swine fever viruses [in Russian]. Veterinariya. 1991;4:28–31.
- Sereda AD, Solovkin S, Phugina L, Makarov V. Immune assays against African swine fever virus [in Russian]. Vopr Virusol. 1992;37:168–70 .PubMedGoogle Scholar
- Sereda AD, Balyshev VM. Antigenic diversity of African swine fever viruses [in Russian]. Vopr Virusol. 2011;56:38–42 .PubMedGoogle Scholar
- Balyshev V, Kurinnov V, Tsibanov S, Kalantaenko Y, Kolbasov D, Pronin V, Biological characteristics of African swine fever virus isolated on the territory of the Russian Federation [in Russian]. Veterinariya. 2010;7:25–7.
- Gallardo C, Mwaengo DM, Macharia JM, Arias M, Taracha EA, Soler A, Enhanced discrimination of African swine fever virus isolates through nucleotide sequencing of the p54, p72, and pB602L (CVR) genes. Virus Genes. 2009;38:85–95. DOIPubMedGoogle Scholar
- Bastos AD, Penrith ML, Crucière C, Edrich JL, Hutchings G, Roger F, Genotyping field strains of African swine fever virus by partial p72 gene characterisation. Arch Virol. 2003;148:693–706. DOIPubMedGoogle Scholar
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–9. DOIPubMedGoogle Scholar
Figures
Table
Cite This Article1These authors contributed equally to this article.
Table of Contents – Volume 21, Number 2—February 2015
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Alexander Malogolovkin, 601120, Vladimirskaya Oblast’, Pokrov, VNIIVViM, Russia
Top