Volume 23, Number 2—February 2017
Dispatch
Determination of Elizabethkingia Diversity by MALDI-TOF Mass Spectrometry and Whole-Genome Sequencing
Figure
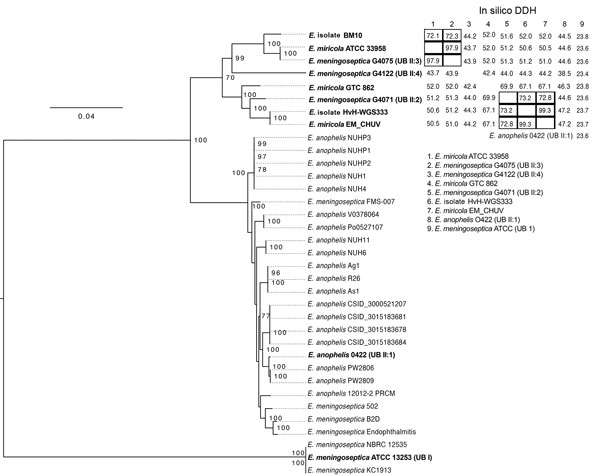
Figure. Phylogenetic tree of Elizabethkingia isolate from a patient with hospital-acquired septic arthritis in Copenhagen, Denmark 2015 compared with reference strains and in silico DNA-DNA hybridization (DDH). Tree was produced by using the Elizabethkingia core genome from all publicly available Elizabethkingia. Bootstrapping support was implemented by running 100 replicates, with values >70% indicated on branches. Initial species identification followed by NCBI isolate name is indicated in the tree. Isolates assigned into Ursing and Bruun (UB) groups and subgroups are indicated in brackets following the isolate name. Table at top right indicates the in silico DDH values; black boxes indicate isolates with in silico DDH >70% indicating they belong to the same species. The E. meningoseptica isolates NBRC 12535, ATCC13253, and KC1913 seem to be identical strains. Scale bar indicates nucleotide substitutions per site.