Volume 24, Number 2—February 2018
CME ACTIVITY - Research
Clinical and Molecular Epidemiology of Staphylococcal Toxic Shock Syndrome in the United Kingdom
Introduction

In support of improving patient care, this activity has been planned and implemented by Medscape, LLC and Emerging Infectious Diseases. Medscape, LLC is jointly accredited by the Accreditation Council for Continuing Medical Education (ACCME), the Accreditation Council for Pharmacy Education (ACPE), and the American Nurses Credentialing Center (ANCC), to provide continuing education for the healthcare team.
Medscape, LLC designates this Journal-based CME activity for a maximum of 1.00 AMA PRA Category 1 Credit(s)™. Physicians should claim only the credit commensurate with the extent of their participation in the activity.
All other clinicians completing this activity will be issued a certificate of participation. To participate in this journal CME activity: (1) review the learning objectives and author disclosures; (2) study the education content; (3) take the post-test with a 75% minimum passing score and complete the evaluation at http://www.medscape.org/journal/eid; and (4) view/print certificate.
Release date: January 16, 2018; Expiration date: January 16, 2019
Learning Objectives
Upon completion of this activity, participants will be able to:
• Assess the clinical features and epidemiology of toxic shock syndrome (TSS) in England, Wales, and Northern Ireland, based on a study using UK national surveillance data
• Identify the molecular epidemiology of TSS in England, Wales, and Northern Ireland
• Discuss superantigen production by dominant TSS strain types and antimicrobial sensitivity of isolates.
CME Editor
Dana C. Dolan, BS, Copyeditor, Emerging Infectious Diseases. Disclosure: Dana C. Dolan, BS, has disclosed no relevant financial relationships.
CME Author
Laurie Barclay, MD, freelance writer and reviewer, Medscape, LLC. Disclosure: Laurie Barclay, MD, has disclosed the following relevant financial relationships: owns stock, stock options, or bonds from Alnylam; Biogen; Pfizer.
Authors
Disclosures: Hema Sharma, PhD, MSc; Debra S. Smith, PhD; Claire E. Turner, PhD; Laurence Game, PhD; Bruno Pichon, PhD; Russell Hope, PhD; Robert Hill, PhD; Angela Kearns, PhD, BSc; and Shiranee Sriskandan, PhD, FRCP, have disclosed no relevant financial relationships.
Abstract
Staphylococcal toxic shock syndrome (TSS) was originally described in menstruating women and linked to TSS toxin 1 (TSST-1)–producing Staphylococcus aureus. Using UK national surveillance data, we ascertained clinical, molecular and superantigenic characteristics of TSS cases. Average annual TSS incidence was 0.07/100,000 population. Patients with nonmenstrual TSS were younger than those with menstrual TSS but had the same mortality rate. Children <16 years of age accounted for 39% of TSS cases, most caused by burns and skin and soft tissue infections. Nonmenstrual TSS is now more common than menstrual TSS in the UK, although both types are strongly associated with the tst+ clonal complex (CC) 30 methicillin-sensitive S. aureus lineage, which accounted for 49.4% of all TSS and produced more TSST-1 and superantigen bioactivity than did tst+ CC30 methicillin-resistant S. aureus strains. Better understanding of this MSSA lineage and infections in children could focus interventions to prevent TSS in the future.
Staphylococcal toxic shock syndrome (TSS) is a life-threatening illness characterized by fever, rash, desquamation, organ dysfunction, and shock. In 1980, the use of highly absorbent tampons in the United States triggered an outbreak of menstrual TSS (mTSS) in young women, and TSS incidence peaked at 13.7/100,000 population (1). Changes in tampon manufacture and advice regarding tampon use helped halt the epidemic. TSS is a notifiable illness in the United States; in 2004–2014, average annual incidence varied from 0.03–0.05/100,000 population (2). In the United Kingdom and other countries in Europe, staphylococcal TSS is not a notifiable illness, so the clinical, microbiological, and toxigenic features of TSS remain poorly described.
TSS is attributed to staphylococcal superantigens that cause massive T-cell activation and cytokine release (3). TSS toxin 1 (TSST-1) is associated with 95% of mTSS cases and 50% of TSS cases caused by nonmenstrual infective foci (nmTSS) (4). Although 24 different staphylococcal superantigens have been described, including staphylococcal enterotoxin (SE) and enterotoxin-like superantigens (5), SE types A, B, and C are implicated in remaining nmTSS cases (3,6), despite the lack of data from Europe.
TSST-1 is encoded by the gene tst, which is carried on mobile genetic elements (MGE) named staphylococcal pathogenicity islands (SaPIs) that lie within the S. aureus chromosome. SaPIs are linked to specific S. aureus genetic families, known as lineages (7). Within human S. aureus strains, tst is carried on SaPI1, SaPI2, and SaP68111 (8,9). Known regulators of tst include the S. aureus accessory gene regulator operon (agr) via the effector molecule RNAIII (10), the staphylococcal respiratory response regulator AB (SrrAB) (10), a glucose catabolite repressor CcpA (11), the staphylococcal accessory regulator A, σB (12) and the SaeRS 2-component system (13).
mTSS strains are reported to belong to a single S. aureus lineage (14,15) corresponding to multilocus sequence type–clonal complex (MLST-CC) 30, a lineage prevalent in the United Kingdom (16). Staphylococcal methicillin resistance is mediated by mecA or mecC genes within the mobile genetic element staphylococcal cassette chromosome mec (SCCmec), of which there are 12 types (17,18). Methicillin-sensitive S. aureus (MSSA) and methicillin-resistant S. aureus (MRSA) strains that are members of CC30 carry tst on SaPI2 (19,20).
In this study, we aimed to characterize the clinical and molecular epidemiology of TSS in England, Wales, and Northern Ireland. We further determined superantigen production by dominant S. aureus strain types.
Case Identification
Public Health England (PHE) requests the referral of all TSS-associated isolates to the national reference laboratory for characterization, including toxin gene profiling. We identified clinician-diagnosed staphylococcal TSS cases from a database of referred S. aureus isolates from England, Wales, and Northern Ireland during January 2008–December 2012 using the search term “toxic shock syndrome.” Clinical and demographic data from the accompanying isolate referral form (Technical Appendix) that had been recorded contemporaneously were scrutinized for accuracy by a clinician (H.S.) before inclusion in the study.
We classified TSS cases in patients <16 years of age as pediatric. We classified cases in female patients 12–60 years of age as mTSS if the infection was associated with menstruation or positive vaginal culture for S. aureus. We classified the remaining cases as nmTSS. All cases had an associated S. aureus isolate.
The average annual incidence of TSS was calculated as cases per 100,000 population using Office for National Statistics UK population estimates (http://www.ons.gov.uk/ons/datasets-and-tables/index.html) and was based on data from 2009 and later (due to changes in reporting practice from November 2008 prompted by national guidance on toxin-producing S. aureus). We used total population for the United Kingdom excluding Scotland as the denominator for all TSS and nmTSS cases; the total female population 12–60 years of age as the denominator for mTSS cases, reflecting the age range of this group; and the number of children <16 years of age as the denominator for pediatric cases. We included data from 2008–2012 in all other analyses.
Molecular Characterization of Isolates
We made MLST-CC assignments on the basis of sequencing the staphylococcal protein A (spa) gene repeat region (21) and referencing spa server (http://spa.ridom.de/mlst.shtml) and MLST (http://saureus.mlst.net) databases. We performed SCCmec detection, typing, and toxin gene profiling (sea-e, seg-j, tst, and pvl only) by multiplex PCR (22,23).
Antimicrobial Susceptibility Testing
For isolates from 2008–2011 (n = 148; Technical Appendix Table 1), we determined antimicrobial MICs by agar dilution (24) and interpreted them in accordance with European Committee on Antimicrobial Susceptibility Testing guidelines (http://www.eucast.org). We did not determine antimicrobial susceptibilities for isolates from 2012.
TSST-1 Production
Based on molecular epidemiologic findings, we assessed TSST-1 production in all tst-positive CC30 MSSA isolates from the TSS cohort (n = 81), including TSS isolates associated with bacteremia, skin and soft tissue infections (SSTI), and deep infections. We also assessed TSST-1 production in randomly selected tst-positive CC30 MRSA isolates from non-TSS patients (n = 39, including carriage, bacteremia, and SSTI isolates) that had been submitted to the reference laboratory during the study period (Technical Appendix Table 1). We quantified TSST-1 in cell-free broth-culture supernatants by Western blot by comparison with purified TSST-1 protein standards (Technical Appendix).
T-Cell Proliferation
We obtained normal-donor peripheral blood mononuclear cells (PBMC) from an approved subcollection of the Imperial College NHS Trust Tissue Bank (ICHTB reference R12023) from anonymized consenting healthy donors. We incubated PBMC (1 × 106 cells/mL) with cell-free RPMI bacterial supernatants (1:1,000 dilution) prepared from tst-positive CC30 MSSA isolates from the TSS cohort (n = 77; 4 of the isolates did not grow in RPMI) and the randomly selected tst-positive CC30 MRSA isolates (n = 39) that were investigated for TSST-1 production. We cultured the PBMC in RPMI medium (Invitrogen, Hemel Hempstead, UK) supplemented with 10% fetal calf serum at 37°C for 48 h in triplicate (25). We measured proliferation after incorporating 1.0 μCi/well of [3H] thymidine and allowing an additional 16 h incubation.
DNA Sequencing and Analysis
We extracted whole genomic DNA from randomly selected tst-positive CC30 MSSA isolates from the TSS cohort (n = 4) and tst-positive CC30 MRSA isolates (n = 5) (Technical Appendix Table 1) (26). We prepared libraries using the Nextera-XT DNA Sample Prep Kit (Illumina, Cambridge, UK) and subjected them to MiSeq sequencing (Illumina), generating 150 bp reads. We deposited data in the GenBank short read archive (accession no. SRP082305). We mapped reads to MLST-CC matched reference genomes MRSA252 (GenBank accession no. NC_002952.2 (27) or MN8 (accession no. NZ_CM000952) using SMALT (http://www.sanger.ac.uk/resources/software/smalt/) and determined single-nucleotide polymorphisms (SNPs) by SAMtools and bcftools (28). We performed de novo assemblies using Velvet (https://www.ebi.ac.uk/~zerbino/velvet/) and annotated them using Prokka (http://www.vicbioinformatics.com/software.prokka.shtml). We used Artemis (http://www.sanger.ac.uk/science/tools/artemis) to visualize the mapping of sequence reads to the reference strain and manually confirm all polymorphisms. For targeted ccpA sequencing, we amplified and sequenced DNA using forward primer 1: 5′- CACAGTGTCGCGTGTTGTTA-3′ and reverse primer 1: 5′- TAAGCGCATCCCTACTGCAC-3′.
Statistical Analysis
We analyzed data with GraphPad Prism 6.0 (GraphPad Software, La Jolla, California, USA). We tested categorical variables using Fisher exact test or χ2 test. We summarized nonparametric data by medians and interquartile ranges (IQR) and compared 2 groups by Mann-Whitney U test. We summarized parametric data by means and SDs and analyzed 2 groups by t-test (2-tailed); we considered p<0.05 significant.
Incidence of TSS
During January 2008–December 2012, a total of 195 TSS case isolates were referred to PHE. We excluded 15 cases from the study (duplicate isolates from the same case, 4 cases; isolates submitted for quality control testing, 2 cases; isolates from cases incorrectly recorded as TSS, 9 cases), leaving 180 microbiologically confirmed TSS cases with isolates. Because of missing clinical data, we were unable to classify 3 isolates as mTSS or nmTSS and could not ascertain the sex of 1 patient with nmTSS.
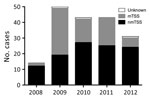
We considered the apparent rise in cases during 2008–2009 an artifact of increased clinical awareness of severe toxigenic S. aureus disease from late 2008, prompted by national guidance on toxin-producing S. aureus (Figure 1). Beginning in 2009, mTSS referrals declined annually, whereas nmTSS cases remained stable. By 2012, cases of nmTSS outnumbered mTSS. Overall, most cases were nonmenstrual (107, 59.4%). Average annual incidence per 100,000 population was 0.07 (95% CI 0.05–0.10) for all cases, 0.09 (95% CI 0.06–0.14) for menstrual cases, and 0.04 (95% CI 0.02–0.06) for nonmenstrual cases.
Clinical Characteristics of TSS Patients
Despite an overall preponderance of female case-patients, we found no gender difference among nmTSS cases (Table 1). The median age of the cohort was 19 years; patients with nmTSS were younger than those with mTSS (median 15.0 vs. 21.5 years; p = 0.01).
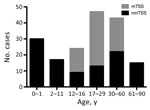
Of the TSS cases studied, 39% (71/180) occurred in children <16 years of age; one sixth of all TSS case-patients were <1 year of age (Figure 2). The median age of pediatric TSS case-patients was 4 years, with an average annual incidence of 0.14/100,000 children (95% CI 0.08–022). However, among children <1 year of age, the average annual incidence increased to 0.45/100,000 (95% CI 0.26–0.79). Most pediatric nmTSS cases were related to burns (26.8%, 15/56) or SSTIs (25%, 14/56).
Five percent of all patients with TSS had died at the time of referral of the isolate. We found no difference in fatality rate between mTSS and nmTSS cases and no association with age (Technical Appendix Table 2). The infective focus in nmTSS cases was SSTI (n = 41), primary bacteremia (n = 15), burns (n = 15), deep abscess (n = 13), respiratory tract (n = 10), bone and joint (n = 4), unknown (n = 6), and other sites (n = 3). We found no association between site of infection and S. aureus lineage (Technical Appendix Figure 1).
Molecular Characteristics of TSS Isolates
Among 180 TSS S. aureus isolates, we identified 88 spa types associated with 15 different MLST-CCs (Technical Appendix Table 3). The leading cause of both mTSS and nmTSS was CC30 MSSA, accounting for >50% of infections (Figure 3), although we found a stronger association of CC30 with mTSS than with nmTSS (72.9% vs. 36.4%; p<0.0001; Technical Appendix Table 3). CC30 MSSA was also the leading cause of TSS among pediatric cases (31/71). We identified only 7 MRSA TSS isolates (Technical Appendix Table 4).
TSS isolates carried 3 superantigen genes on average (Technical Appendix Table 5). The most common superantigen gene among both mTSS and nmTSS isolates was tst (Table 2; Technical Appendix Figure 2), with the exception of the other 2 prevalent superantigen genes, seg and sei, that are carried on an enterotoxin gene cluster (egc) along with selm/n/o/u in most S. aureus isolates (5). The tst gene was associated with mTSS (Table 2) and strongly associated with the CC30 lineage of S. aureus (Technical Appendix Table 6). The superantigen gene sea combined with tst was also linked to mTSS (Table 2), whereas sea alone was associated with CC30 (Technical Appendix Tables 5, 6); sec was linked to nmTSS (Table 2) and CC45 (Technical Appendix Table 5). Ten nmTSS cases were associated with isolates that lacked any superantigen gene tested; 7 were CC15, highlighting severe disease attributable to this lineage that was unexplained by the presence of major superantigens (Technical Appendix Tables 5, 6).
Antimicrobial Susceptibility of TSS Isolates
Most isolates were MSSA (mecA negative). The rate of resistance to erythromycin was 9.2%; to ciprofloxacin, 8.5%; to tetracycline, 3.5%; and to teicoplanin, 1.4%. For 7 mecA-positive MRSA-TSS isolates, the resistance rate to ciprofloxacin was 57.1%; to erythromycin, 42.6%; and to clindamycin, 14.3%.
As MRSA-related TSS is rarely reported we examined these cases in more detail. All 7 MRSA cases were nonmenstrual, affecting mainly male patients; 3 were associated with SSTIs. The median patient age was 34 (IQR 2.3–64.3) years. Five isolates were identified as CC22-SCCmecIV, and 4 carried sec, corresponding to the healthcare-associated MRSA clade dominant in the UK, EMRSA-15; MRSA-TSS cases showed a clear association with this lineage (Technical Appendix Table 4). The remaining CC22 isolate carried tst and belonged to a MRSA lineage frequently identified in the Middle East. Only 1 MRSA-TSS isolate was CC30-SCCmecII, corresponding to the UK HA-MRSA clade EMRSA-16. One isolate was CC6-SCCmecII and lacked all superantigen genes tested.
TSST-1 Production by CC30 S. aureus
The strong association of CC30 with TSS was unsurprising because of the presence of tst. We measured TSST-1 in broth-culture supernatants from tst-positive CC30 MSSA isolates from the TSS cohort and, for comparison, randomly selected clinical tst-positive MRSA isolates that belonged to the same lineage (CC30) (20,29) (Technical Appendix Table 1).
Of note, 77/81 tst-positive CC30 MSSA isolates produced detectable TSST-1, compared with 9/39 tst-positive CC30 MRSA isolates. The tst-positive CC30 MSSA isolates produced more TSST-1 than did tst-positive CC30 MRSA isolates, albeit with marked variability (88.5 ± 48.3 vs. 31.4 ± 18.1 ng/mL,;p<0.0001; Figure 4, panel A). Furthermore, the superantigenic activity of isolates, measured by T-cell proliferation in response to broth-culture supernatants, of tst-positive CC30 MSSA strains (164,893 ± 36,191 counts/min) was significantly greater than that of tst-positive CC30 MRSA strains (149,653 ± 30,412 counts/min; p = 0.02; Figure 4, panel B).
tst-positive CC30 MRSA and Mutation in tst Regulator, CcpA
To ascertain the basis for the observed variability in TSST-1 production among CC30 S. aureus, we subjected 4 tst-positive CC30 MSSA isolates from the TSS cohort and 5 tst-positive CC30 MRSA clinical isolates to whole-genome sequencing. The tst gene, promoter, and regulator sequences, including SarA, SrrAB, agr, and σB, were identical among the 9 sequenced strains and reference isolates (MN8/MRSA252).
We detected mutations in TSST-1 regulator SaeRS in 2/4 tst-positive CC30 MSSA isolates; a synonymous SNP C481T in SaeR in 1 strain and a nonsynonymous SNP in SaeS in another resulted in a change from asparagine to serine at aa residue 218. Because these strains produced abundant TSST-1 (Technical Appendix Table 1), we did not study these mutations further.
We detected a nonsynonymous SNP in the sequence of regulator ccpA in all 5 tst-positive CC30 MRSA isolates but not in any tst-positive CC30 MSSA isolate. This difference translated into a change from threonine (ACA) to isoleucine (ATA) at aa residue 87/329 (Technical Appendix Figure 3).
To determine the prevalence of the ccpA (T87I) variant in CC30, we sequenced ccpA in an additional 34 tst-positive CC30 MRSA and 19 tst-positive CC30 MSSA isolates (Technical Appendix Table 1). Including genome-sequenced isolates, 33/39 tst-positive CC30 MRSA isolates had ccpA (T87I), compared with 0/23 tst-positive CC30 MSSA isolates, confirming an association of ccpA (T87I) with CC30 MRSA strains. Furthermore, ccpA (T87I) was strongly negatively associated with production of TSST-1 in tst-positive CC30 S. aureus: 26/33 ccpA (T87I) isolates did not produce TSST-1, compared with only 1/23 wild-type ccpA isolates (p<0.0001 by Fisher exact test).
We conducted SCCmec typing on a subset of tst-positive CC30 MRSA strains (n = 15; Technical Appendix Table 1). Results demonstrated an association of ccpA (T87I) with SCCmecII; 7/11 SCCmecII isolates had ccpA (T87I), compared with 0/4 SCCmecIV isolates (p = 0.03 by χ2 test). This finding highlights the possibility that reduced TSST-1 production might be attributable to either SCCmecII or ccpA (T87I).
We provide a substantive national clinical and microbiological overview of staphylococcal TSS cases in the United Kingdom. TSS incidence was 0.07/100,000 population, nmTSS cases now outnumber mTSS cases, and nmTSS affects younger persons. The tst-positive CC30 S. aureus lineage was linked strongly with TSS and almost all mTSS cases. CC30 MSSA is a prevalent lineage in the United Kingdom (16), so ongoing surveillance and clinical vigilance for TSS are important.
Our findings may underestimate TSS incidence because notification of TSS is voluntary in the United Kingdom and we included only microbiologically confirmed cases. These factors increase diagnostic confidence, but TSS is a syndromic condition not requiring bacteriological confirmation. Overall TSS incidence was low but similar to rates in the United States (2); improvements in care may account for low overall incidence of TSS, because patients may not fulfill all of the criteria required by the case definition of TSS. The overall TSS incidence in children contrasts with findings from a British Pediatric Surveillance Unit study in which a higher incidence of combined streptococcal and staphylococcal TSS cases was reported (30).
The number of cases of mTSS fell from 2009 to 2012, such that nmTSS cases are now more common than mTSS cases, mirroring US trends (31). Patients in our study were younger than in US cohorts (31,32), and nmTSS patients were younger than those with mTSS. Most nmTSS cases occurred in children, with burns and SSTIs as the cause in 51.8% (29/56) of these cases. An association between nmTSS and increased mortality rate has been reported, although a high incidence of bacteremia may have affected the findings of that study (33). It is possible that we did not ascertain all cases of TSS, although we found no difference in reported deaths between mTSS and nmTSS cases or associations with age; the overall death rate was 5%.
The association of TSS, and particularly mTSS, with a single lineage corresponding to CC30 S. aureus has been described in diverse geographic localities (14,15). The tst-positive CC30 MSSA clone has recently been named epidemic MSSA-ST30 because it is responsible for a substantial amount of S. aureus disease and is a precursor to the HA-MRSA clone, EMRSA-16, which has been responsible for major national UK MRSA outbreaks (29).
The tst gene was the predominant superantigen gene among TSS isolates, excluding seg and sei, which were also previously implicated in TSS (34). The superantigens seg and sei are carried on the egc, which is widespread in S. aureus (5), and are unlikely to have any specific association with TSS. We linked tst to mTSS and CC30. Several groups have demonstrated similar associations of staphylococcal superantigen genes with specific lineages (35,36), due to clonal associations, superantigen arrangements, and transmission via mobile genetic elements, although other firm associations linking lineage, superantigen gene carriage, infection type, and disease presentations have not been made. A recent study of atopic dermatitis that examined the relationship of ethnicity and staphylococcal virulence factors found a lack of tst-positive S. aureus atopic dermatitis in African American persons that was consistent with an absence of tst-positive S. aureus mTSS among this group, suggesting differences in disease presentation among disparate ethnic groups (37) based on host characteristics. The ethnicity of the patients with TSS referred to PHE in this study was not recorded, and such bacterial genetic associations with disease could not be made but may merit consideration in future studies.
Among MSSA isolates, resistance rates to key antimicrobial drugs were similar to reported UK MSSA bacteremia isolates (38). Notably, teicoplanin resistance was detected, although rarely. This finding circumvents any need to change current recommendations for antimicrobial drugs for TSS that include a bactericidal cell wall inhibitor (e.g., β-lactamase–resistant antistaphylococcal) and protein-synthesis inhibitor (e.g., clindamycin) along with intravenous immunoglobulin for severe cases unresponsive to first-line therapy and source control (39). No vaccines are available to prevent TSS, although a recombinant TSST-1 variant vaccine has shown promise in a recent human clinical trial and was found to be safe and immunogenic (40).
The MRSA-TSS rate in this study was lower than rates in the United States (32), perhaps reflecting the low UK community-associated MRSA prevalence (41). All MRSA cases were nonmenstrual and mostly associated with recognized healthcare-associated MRSA clones, although we did not record the mode of acquisition. Only 1 CC30 MRSA (EMRSA-16) isolate caused TSS, even though CC30 is the main TSS-associated lineage; this finding mirrors the national decline in UK EMRSA-16 over time (42).
Isolates of tst-positive CC30 MSSA were more likely to produce TSST-1 in vitro and secreted almost 3 times more TSST-1 than did tst-positive CC30 MRSA isolates, which translated into a functional difference in superantigenic activity. We do not know whether such a difference would extend to the in vivo setting. Our study of TSST-1 production was limited by availability of clinical tst+ CC30 strains; clinical TSS CC30 MSSA strains were therefore compared with clinical non-TSS CC30 MRSA strains and not to clinical TSS MRSA strains. Thus, more MSSA than MRSA strains were from the genital tract or from burns, potentially confounding phenotypic differences observed. Defining the precise comparator group for TSS CC30 MSSA isolates is challenging because of lack of TSS CC30 MRSA isolates and suitable non-TSS strains referred to PHE.
Bacterial acquisition of antimicrobial drug resistance elements can be associated with a fitness cost. In the United Kingdom, most CC30 HA-MRSA strains carry SCCmecII (EMRSA-16; ST36-SCCmecII) that may reduce cytolytic toxin production and, in association with fudoh gene carriage by this element, reduce hemolytic activity and virulence (43,44). Our findings suggest an association between SCCmecII and reduced TSST-1 production that might be linked to a SNP in a regulatory gene, ccpA. The resulting mutation in CcpA occurs adjacent to a co-repressor binding site in the transcriptional regulation region (Technical Appendix Figure 3) that could influence tst promoter binding and affect TSST-1 secretion. Such SNPs in virulence regulators may have had a role in shaping the healthcare-associated phenotype of EMRSA-16 (20). New tools that allow manipulation of previously nontransformable lineages such as CC30 will facilitate investigating such genetic mechanisms in S. aureus (45).
Our study shows that the ability to produce TSST-1 varies widely within the tst-positive CC30 lineage and impaired expression is associated with the presence of SCCmecII and ccpA (T87I), underlining the potential for genomic approaches to contribute to greater understanding of patterns of clinical disease. Given the prevalence of tst-positive CC30 MSSA causing TSS and its role as a dominant UK lineage of S. aureus, active surveillance of this lineage is required. Clarification of the particular modes of transmission, acquisition, and pathogenesis of this lineage may identify susceptible persons, such as younger persons with burns and SSTIs, who might benefit from interventions such as vaccination with recombinant TSST-1 or S. aureus screening and decolonization in the future to prevent the occurrence of this life-threatening syndrome.
Dr. Sharma is a physician in infectious diseases, microbiology, and virology and a clinical research fellow at Imperial College London. Her primary research interests relate to the pathogenesis of staphylococcal disease.
Acknowledgments
The authors acknowledge the NIHR Biomedical Research Centre awarded to Imperial College Healthcare NHS Trust and the Imperial College Healthcare NHS Trust Tissue Bank.
This work was funded by the UK Clinical Research Collaboration (UKCRC, Research Training Fellowship G0800777/1 to H.S., and the Centre for Infection Prevention and Management) and by the National Institute for Health Research (NIHR) Health Protection Research Unit in Healthcare-associated Infections and Antimicrobial Resistance at Imperial College London in partnership with Public Health England. The views expressed are those of the authors and not necessarily those of the National Health Service, the NIHR, the Department of Health or Public Health England.
References
- Osterholm MT, Forfang JC. Toxic-shock syndrome in Minnesota: results of an active-passive surveillance system. J Infect Dis. 1982;145:458–64. DOIPubMedGoogle Scholar
- Adams DA, Thomas KR, Jajosky RA, Foster L, Sharp P, Onweh DH, et al.; Nationally Notifiable Infectious Conditions Group. Summary of notifiable infectious diseases and conditions—United States, 2014. MMWR Morb Mortal Wkly Rep. 2016;63:1–152. DOIPubMedGoogle Scholar
- Fraser JD, Proft T. The bacterial superantigen and superantigen-like proteins. Immunol Rev. 2008;225:226–43. DOIPubMedGoogle Scholar
- Bohach GA, Fast DJ, Nelson RD, Schlievert PM. Staphylococcal and streptococcal pyrogenic toxins involved in toxic shock syndrome and related illnesses. Crit Rev Microbiol. 1990;17:251–72. DOIPubMedGoogle Scholar
- Grumann D, Nübel U, Bröker BM. Staphylococcus aureus toxins—their functions and genetics. Infect Genet Evol. 2014;21:583–92. DOIPubMedGoogle Scholar
- Whiting JL, Rosten PM, Chow AW. Determination by western blot (immunoblot) of seroconversions to toxic shock syndrome (TSS) toxin 1 and enterotoxin A, B, or C during infection with TSS- and non-TSS-associated Staphylococcus aureus. Infect Immun. 1989;57:231–4.PubMedGoogle Scholar
- Novick RP. Mobile genetic elements and bacterial toxinoses: the superantigen-encoding pathogenicity islands of Staphylococcus aureus. Plasmid. 2003;49:93–105. DOIPubMedGoogle Scholar
- Novick RP, Christie GE, Penadés JR. The phage-related chromosomal islands of Gram-positive bacteria. Nat Rev Microbiol. 2010;8:541–51. DOIPubMedGoogle Scholar
- Li Z, Stevens DL, Hamilton SM, Parimon T, Ma Y, Kearns AM, et al. Fatal S. aureus hemorrhagic pneumonia: genetic analysis of a unique clinical isolate producing both PVL and TSST-1. PLoS One. 2011;6:e27246. DOIPubMedGoogle Scholar
- Pragman AA, Schlievert PM. Virulence regulation in Staphylococcus aureus: the need for in vivo analysis of virulence factor regulation. FEMS Immunol Med Microbiol. 2004;42:147–54. DOIPubMedGoogle Scholar
- Seidl K, Bischoff M, Berger-Bächi B. CcpA mediates the catabolite repression of tst in Staphylococcus aureus. Infect Immun. 2008;76:5093–9. DOIPubMedGoogle Scholar
- Andrey DO, Jousselin A, Villanueva M, Renzoni A, Monod A, Barras C, et al. Impact of the regulators sigB, rot, sarA and sarS on the toxic shock tst promoter and TSST-1 expression in Staphylococcus aureus. PLoS One. 2015;10:e0135579. DOIPubMedGoogle Scholar
- Baroja ML, Herfst CA, Kasper KJ, Xu SX, Gillett DA, Li J, et al. The SaeRS two-component system is a direct and dominant transcriptional activator of toxic shock syndrome toxin 1 in Staphylococcus aureus. J Bacteriol. 2016;198:2732–42. DOIPubMedGoogle Scholar
- Musser JM, Schlievert PM, Chow AW, Ewan P, Kreiswirth BN, Rosdahl VT, et al. A single clone of Staphylococcus aureus causes the majority of cases of toxic shock syndrome. Proc Natl Acad Sci U S A. 1990;87:225–9. DOIPubMedGoogle Scholar
- Fitzgerald JR, Sturdevant DE, Mackie SM, Gill SR, Musser JM. Evolutionary genomics of Staphylococcus aureus: insights into the origin of methicillin-resistant strains and the toxic shock syndrome epidemic. Proc Natl Acad Sci U S A. 2001;98:8821–6. DOIPubMedGoogle Scholar
- Feil EJ, Cooper JE, Grundmann H, Robinson DA, Enright MC, Berendt T, et al. How clonal is Staphylococcus aureus? J Bacteriol. 2003;185:3307–16. DOIPubMedGoogle Scholar
- Hiramatsu K, Ito T, Tsubakishita S, Sasaki T, Takeuchi F, Morimoto Y, et al. Genomic basis for methicillin resistance in Staphylococcus aureus. Infect Chemother. 2013;45:117–36. DOIPubMedGoogle Scholar
- Wu Z, Li F, Liu D, Xue H, Zhao X. Novel type XII staphylococcal cassette chromosome mec harboring a new cassette chromosome recombinase, CcrC2. Antimicrob Agents Chemother. 2015;59:7597–601. DOIPubMedGoogle Scholar
- Subedi A, Ubeda C, Adhikari RP, Penadés JR, Novick RP. Sequence analysis reveals genetic exchanges and intraspecific spread of SaPI2, a pathogenicity island involved in menstrual toxic shock. Microbiology. 2007;153:3235–45. DOIPubMedGoogle Scholar
- McAdam PR, Templeton KE, Edwards GF, Holden MT, Feil EJ, Aanensen DM, et al. Molecular tracing of the emergence, adaptation, and transmission of hospital-associated methicillin-resistant Staphylococcus aureus. Proc Natl Acad Sci U S A. 2012;109:9107–12. DOIPubMedGoogle Scholar
- Koreen L, Ramaswamy SV, Graviss EA, Naidich S, Musser JM, Kreiswirth BN. spa typing method for discriminating among Staphylococcus aureus isolates: implications for use of a single marker to detect genetic micro- and macrovariation. J Clin Microbiol. 2004;42:792–9. DOIPubMedGoogle Scholar
- Milheiriço C, Oliveira DC, de Lencastre H. Update to the multiplex PCR strategy for assignment of mec element types in Staphylococcus aureus. Antimicrob Agents Chemother. 2007;51:3374–7. DOIPubMedGoogle Scholar
- Boakes E, Kearns AM, Ganner M, Perry C, Warner M, Hill RL, et al. Molecular diversity within clonal complex 22 methicillin-resistant Staphylococcus aureus encoding Panton-Valentine leukocidin in England and Wales. Clin Microbiol Infect. 2011;17:140–5. DOIPubMedGoogle Scholar
- Andrews JM. Determination of minimum inhibitory concentrations. J Antimicrob Chemother. 2001;48(Suppl 1):5–16. DOIPubMedGoogle Scholar
- Unnikrishnan M, Altmann DM, Proft T, Wahid F, Cohen J, Fraser JD, et al. The bacterial superantigen streptococcal mitogenic exotoxin Z is the major immunoactive agent of Streptococcus pyogenes. J Immunol. 2002;169:2561–9. DOIPubMedGoogle Scholar
- Pospiech A, Neumann B. A versatile quick-prep of genomic DNA from gram-positive bacteria. Trends Genet. 1995;11:217–8. DOIPubMedGoogle Scholar
- Holden MT, Feil EJ, Lindsay JA, Peacock SJ, Day NP, Enright MC, et al. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc Natl Acad Sci U S A. 2004;101:9786–91. DOIPubMedGoogle Scholar
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. DOIPubMedGoogle Scholar
- Aanensen DM, Feil EJ, Holden MT, Dordel J, Yeats CA, Fedosejev A, et al.; European SRL Working Group. Whole-genome sequencing for routine pathogen surveillance in public health: a population snapshot of invasive Staphylococcus aureus in Europe. MBio. 2016;7:e00444–16. DOIPubMedGoogle Scholar
- Adalat S, Dawson T, Hackett SJ, Clark JE; In association with the British Paediatric Surveillance Unit. Toxic shock syndrome surveillance in UK children. Arch Dis Child. 2014;99:1078–82. DOIPubMedGoogle Scholar
- Hajjeh RA, Reingold A, Weil A, Shutt K, Schuchat A, Perkins BA. Toxic shock syndrome in the United States: surveillance update, 1979 1996. Emerg Infect Dis. 1999;5:807–10. DOIPubMedGoogle Scholar
- DeVries AS, Lesher L, Schlievert PM, Rogers T, Villaume LG, Danila R, et al. Staphylococcal toxic shock syndrome 2000-2006: epidemiology, clinical features, and molecular characteristics. PLoS One. 2011;6:e22997. DOIPubMedGoogle Scholar
- Descloux E, Perpoint T, Ferry T, Lina G, Bes M, Vandenesch F, et al. One in five mortality in non-menstrual toxic shock syndrome versus no mortality in menstrual cases in a balanced French series of 55 cases. Eur J Clin Microbiol Infect Dis. 2008;27:37–43. DOIPubMedGoogle Scholar
- Jarraud S, Cozon G, Vandenesch F, Bes M, Etienne J, Lina G. Involvement of enterotoxins G and I in staphylococcal toxic shock syndrome and staphylococcal scarlet fever. J Clin Microbiol. 1999;37:2446–9.PubMedGoogle Scholar
- Holtfreter S, Grumann D, Schmudde M, Nguyen HT, Eichler P, Strommenger B, et al. Clonal distribution of superantigen genes in clinical Staphylococcus aureus isolates. J Clin Microbiol. 2007;45:2669–80. DOIPubMedGoogle Scholar
- Jarraud S, Mougel C, Thioulouse J, Lina G, Meugnier H, Forey F, et al. Relationships between Staphylococcus aureus genetic background, virulence factors, agr groups (alleles), and human disease. Infect Immun. 2002;70:631–41. DOIPubMedGoogle Scholar
- Merriman JA, Mueller EA, Cahill MP, Beck LA, Paller AS, Hanifin JM, et al. Temporal and racial differences associated with atopic dermatitis Staphylococcus aureus and encoded virulence factors. mSphere. 2016;1:e00295-16.
- Public Health England. Voluntary reporting of Staphylococcus aureus bacteraemia in England, Wales, and Northern Ireland, 2013 [cited 2015 Mar 6]. http://www.gov.uk/government/uploads/system/uploads/attachment_data/file/346324/Voluntary_reporting_S._aureus_bacteraemia_England_Wales_Northern_Ireland_2013.pdf.
- American Academy of Pediatrics. Staphylococcal infections. In: Kimberlin DW, Brady MT, Jackson MA, Long SS. Red book: 2015 report of the Committee on Infectious Diseases. Elk Grove Village (IL): American Academy of Pediatrics, 2015. p. 715–32.
- Schwameis M, Roppenser B, Firbas C, Gruener CS, Model N, Stich N, et al. Safety, tolerability, and immunogenicity of a recombinant toxic shock syndrome toxin (rTSST)-1 variant vaccine: a randomised, double-blind, adjuvant-controlled, dose escalation first-in-man trial. Lancet Infect Dis. 2016;16:1036–44. DOIPubMedGoogle Scholar
- Elston JW, Barlow GD. Community-associated MRSA in the United Kingdom. J Infect. 2009;59:149–55. DOIPubMedGoogle Scholar
- Ellington MJ, Hope R, Livermore DM, Kearns AM, Henderson K, Cookson BD, et al. Decline of EMRSA-16 amongst methicillin-resistant Staphylococcus aureus causing bacteraemias in the UK between 2001 and 2007. J Antimicrob Chemother. 2010;65:446–8. DOIPubMedGoogle Scholar
- Rudkin JK, Edwards AM, Bowden MG, Brown EL, Pozzi C, Waters EM, et al. Methicillin resistance reduces the virulence of healthcare-associated methicillin-resistant Staphylococcus aureus by interfering with the agr quorum sensing system. J Infect Dis. 2012;205:798–806. DOIPubMedGoogle Scholar
- Kaito C, Omae Y, Matsumoto Y, Nagata M, Yamaguchi H, Aoto T, et al. A novel gene, fudoh, in the SCCmec region suppresses the colony spreading ability and virulence of Staphylococcus aureus. PLoS One. 2008;3:e3921. DOIPubMedGoogle Scholar
- Monk IR, Shah IM, Xu M, Tan MW, Foster TJ. Transforming the untransformable: application of direct transformation to manipulate genetically Staphylococcus aureus and Staphylococcus epidermidis. MBio. 2012;3:e00277–11. DOIPubMedGoogle Scholar
Figures
Tables
Follow Up
Earning CME Credit
To obtain credit, you should first read the journal article. After reading the article, you should be able to answer the following, related, multiple-choice questions. To complete the questions (with a minimum 75% passing score) and earn continuing medical education (CME) credit, please go to http://www.medscape.org/journal/eid. Credit cannot be obtained for tests completed on paper, although you may use the worksheet below to keep a record of your answers.
You must be a registered user on http://www.medscape.org. If you are not registered on http://www.medscape.org, please click on the “Register” link on the right hand side of the website.
Only one answer is correct for each question. Once you successfully answer all post-test questions, you will be able to view and/or print your certificate. For questions regarding this activity, contact the accredited provider, CME@medscape.net. For technical assistance, contact CME@medscape.net. American Medical Association’s Physician’s Recognition Award (AMA PRA) credits are accepted in the US as evidence of participation in CME activities. For further information on this award, please go to https://www.ama-assn.org. The AMA has determined that physicians not licensed in the US who participate in this CME activity are eligible for AMA PRA Category 1 Credits™. Through agreements that the AMA has made with agencies in some countries, AMA PRA credit may be acceptable as evidence of participation in CME activities. If you are not licensed in the US, please complete the questions online, print the AMA PRA CME credit certificate, and present it to your national medical association for review.
Article Title:
Clinical and Molecular Epidemiology of Staphylococcal Toxic Shock Syndrome in the United Kingdom
CME Questions
1. Your patient is a 16-year-old girl with toxic shock syndrome (TSS). According to the study using UK national surveillance data by Sharma and colleagues, which of the following statements about the clinical features and epidemiology of TSS in England, Wales, and Northern Ireland is correct?
A. Menstrual TSS (mTSS) is more prevalent than nonmenstrual TSS (nmTSS)
B. Patients with nmTSS were older than those with mTSS and had higher mortality rates
C. An estimated 39% of TSS cases occurred in children age 16 years or younger, mostly caused by burns and skin and soft tissue infections
D. This study most likely overestimated the incidence of TSS
2. In the study using UK national surveillance data by Sharma and colleagues, which of the following statements about the molecular epidemiology of TSS in England, Wales, and Northern Ireland is correct?
A. Toxic shock syndrome toxin 1 (TSST-1)-producing (tst+) CC30 lineage methicillin-sensitive Staphylococcus aureus (MSSA) strains were the leading cause of mTSS but not nmTSS
B. Improved understanding of the tst + CC30 MSSA lineage could help focus future preventive interventions in children (eg, vaccination with recombinant TSST-1 or S aureus screening and decolonization)
C. The MSSA-ST30 clone is uncommon in the United Kingdom
D. CC30 MSSA was not isolated from pediatric cases of TSS
3. According to the study using UK national surveillance data by Sharma and colleagues, which of the following statements about superantigen production by dominant TSS strain types and antimicrobial sensitivity of isolates is correct?
A. tst + CC30 lineage MSSA produced more TSST-1 (88.5 ±48.3 vs. 31.4 ±18.1 ng/mL; p<0.0001) and superantigen bioactivity than tst + CC30 MRSA strains
B. Most isolates were resistant to methicillin
C. The findings call for a change in current antimicrobial recommendations for TSS
D. tst was the only superantigen gene among TSS isolates
1Current affiliation: University of Sheffield, Sheffield, UK.
2These authors contributed equally to this article.
Related Links
Table of Contents – Volume 24, Number 2—February 2018
| EID Search Options |
|---|
|
|
|
|
|
|


Please use the form below to submit correspondence to the authors or contact them at the following address:
Shiranee Sriskandan, Imperial College London, Department of Medicine, Hammersmith Campus, Du Cane Road, London W12 0NN, UK
Top