Volume 25, Number 10—October 2019
Dispatch
Genetic Characterization and Zoonotic Potential of Highly Pathogenic Avian Influenza Virus A(H5N6/H5N5), Germany, 2017–2018
Figure
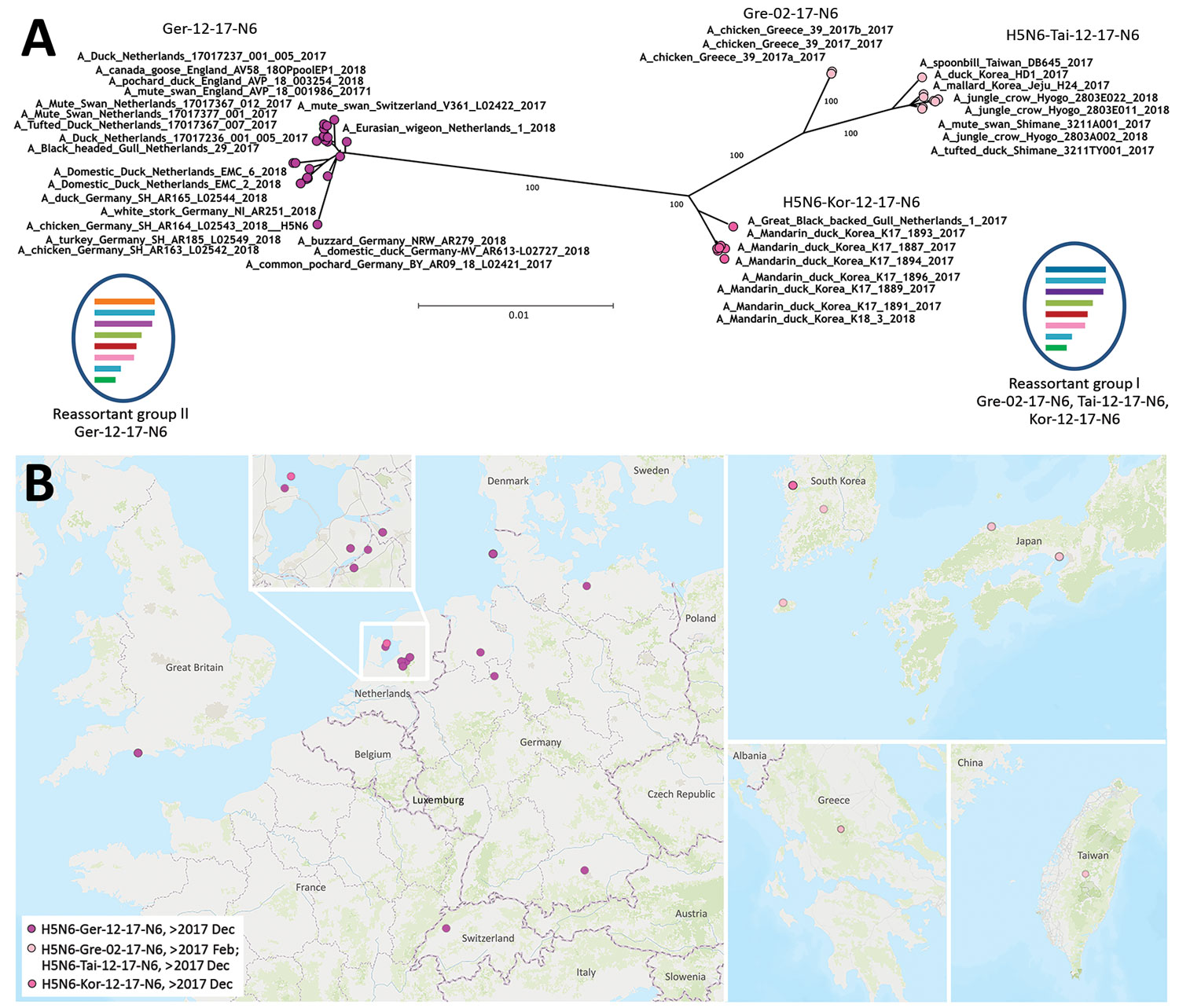
Figure. Phylogenetic clustering and geographic distribution of highly pathogenic avian influenza A(H5N6) viruses, Europe, 2017–2018. A) Supernetwork generated by using maximum-likelihood trees of influenza virus full genomes with RAxML (https://cme.h-its.org/exelixis/web/software/raxml/index.html) and 1,000 bootstrap iterations followed by network analysis with SplitsTree4 (http://ab.inf.uni-tuebingen.de/software/splitstree4). Reassortant viruses are grouped according to their phylogenetic results. Scale bar indicates nucleotide substitutions per site. The mosaic genome structure of reassortant groups I and II is also provided. Gene segment descriptions are given in Appendix
1These authors contributed equally to this article.
Page created: September 17, 2019
Page updated: September 17, 2019
Page reviewed: September 17, 2019
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.