Volume 25, Number 2—February 2019
Dispatch
Bat Influenza A(HL18NL11) Virus in Fruit Bats, Brazil
Figure 1
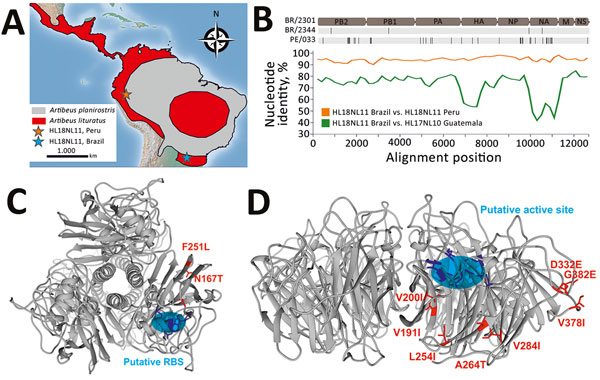
Figure 1. Bat influenza A(HL18NL11) virus detection and genomic characterization, Brazil, 2010–2014. A) Distribution of Artibeus species bats carrying HL18NL11 in Central and South America, according to the Red List of Threatened Species from the International Union for Conservation of Nature (https://www.iucnredlist.org). Orange star indicates the sampling site of an HL18NL11-positive bat in Peru (5); blue star indicates the sampling site of the HL18NL11-positive bats in Brazil for this study. Maps were created using QGIS2.14.3 (http://www.qgis.org) with data freely available from http://www.naturalearthdata.com. B) Top, schematic representation of the genome organization of A/great fruit-eating bat/Brazil/2301/2012 (HL18NL11) and amino acid exchanges (black lines) compared with A/great fruit-eating bat/Brazil/2344/2012 (HL18NL11) and Peru HL18NL11 (GenBank accession nos. CY125942–49). Nucleotide sequence identities between the concatenated HL18NL11 (Brazil), HL17NL10, and HL18NL11 (Guatemala and Peru) sequences were calculated in SSE version 1.2 (http://www.virus-evolution.org/Downloads/Software) with a sliding window of 200 and step size of 100 nt. C) Homology model of the HL protein of A/great fruit-eating bat/Brazil/2301/2012 viewed from the top, modeled on the published crystal structure retrieved from the SWISS-MODEL repository (https://www.swissmodel.expasy.org). The putative RBS is shown in blue, 3 highly conserved residues (W153, H183, and Y195) in HAs and HLs are in purple, and amino acid substitutions between Brazil strains and the Peru prototype strain are in red. D) Homology model of the NL of A/great fruit-eating bat/Brazil/2301/2012 viewed from the top, constructed as in panel C. The putative active site is shown in a blue circle, the 6 residues (R118, W178, S179, R224, E276 and E425) conserved in influenza A virus neuraminidase genes are in purple, and amino acid substitutions between Brazil strains and the Peru prototype strain are in red. HA, hemagglutinin; HL, HA-like; NL, neuraminidase-like; RBS, receptor-binding site.
References
- Olsen B, Munster VJ, Wallensten A, Waldenström J, Osterhaus ADME, Fouchier RAM. Global patterns of influenza a virus in wild birds. Science. 2006;312:384–8. DOIPubMedGoogle Scholar
- Brunotte L, Beer M, Horie M, Schwemmle M. Chiropteran influenza viruses: flu from bats or a relic from the past? Curr Opin Virol. 2016;16:114–9. DOIPubMedGoogle Scholar
- Olival KJ, Hosseini PR, Zambrana-Torrelio C, Ross N, Bogich TL, Daszak P. Host and viral traits predict zoonotic spillover from mammals. Nature. 2017;546:646–50. DOIPubMedGoogle Scholar
- Tong S, Li Y, Rivailler P, Conrardy C, Castillo DA, Chen LM, et al. A distinct lineage of influenza A virus from bats. Proc Natl Acad Sci U S A. 2012;109:4269–74. DOIPubMedGoogle Scholar
- Tong S, Zhu X, Li Y, Shi M, Zhang J, Bourgeois M, et al. New world bats harbor diverse influenza A viruses. PLoS Pathog. 2013;9:e1003657. DOIPubMedGoogle Scholar
- García-Sastre A. The neuraminidase of bat influenza viruses is not a neuraminidase. Proc Natl Acad Sci U S A. 2012;109:18635–6. DOIPubMedGoogle Scholar
- Góes LGB, Campos ACA, Carvalho C, Ambar G, Queiroz LH, Cruz-Neto AP, et al. Genetic diversity of bats coronaviruses in the Atlantic Forest hotspot biome, Brazil. Infect Genet Evol. 2016;44:510–3. DOIPubMedGoogle Scholar
- Anthony SJ, Islam A, Johnson C, Navarrete-Macias I, Liang E, Jain K, et al. Non-random patterns in viral diversity. Nat Commun. 2015;6:8147. DOIPubMedGoogle Scholar
- Drexler JF, Corman VM, Wegner T, Tateno AF, Zerbinati RM, Gloza-Rausch F, et al. Amplification of emerging viruses in a bat colony. Emerg Infect Dis. 2011;17:449–56. DOIPubMedGoogle Scholar
- Zhu X, Yu W, McBride R, Li Y, Chen LM, Donis RO, et al. Hemagglutinin homologue from H17N10 bat influenza virus exhibits divergent receptor-binding and pH-dependent fusion activities. Proc Natl Acad Sci U S A. 2013;110:1458–63. DOIPubMedGoogle Scholar
- Rejmanek D, Hosseini PR, Mazet JAK, Daszak P, Goldstein T. Evolutionary dynamics and global diversity of influenza A virus. J Virol. 2015;89:10993–1001. DOIPubMedGoogle Scholar
- Larsen PA, Marchán-Rivadeneira MR, Baker RW. Speciation dynamics of the fruit-eating bats (genus Artibeus): with evidence of ecological divergence in Central American populations. In: Adams RA, Pedersen SC, editors. Bat evolution, ecology, and conservation. New York: Springer Science + Business Media; 2013. p. 315–339.
- Drexler JF, Corman VM, Drosten C. Ecology, evolution and classification of bat coronaviruses in the aftermath of SARS. Antiviral Res. 2014;101:45–56. DOIPubMedGoogle Scholar
- Moreira ÉA, Locher S, Kolesnikova L, Bolte H, Aydillo T, García-Sastre A, et al. Synthetically derived bat influenza A-like viruses reveal a cell type- but not species-specific tropism. Proc Natl Acad Sci U S A. 2016;113:12797–802. DOIPubMedGoogle Scholar
- Munster VJ, Adney DR, van Doremalen N, Brown VR, Miazgowicz KL, Milne-Price S, et al. Replication and shedding of MERS-CoV in Jamaican fruit bats (Artibeus jamaicensis). Sci Rep. 2016;6:21878. DOIPubMedGoogle Scholar
1These senior authors contributed equally to this article.