Volume 25, Number 7—July 2019
Research
Nationwide Stepwise Emergence and Evolution of Multidrug-Resistant Campylobacter jejuni Sequence Type 5136, United Kingdom
Figure 4
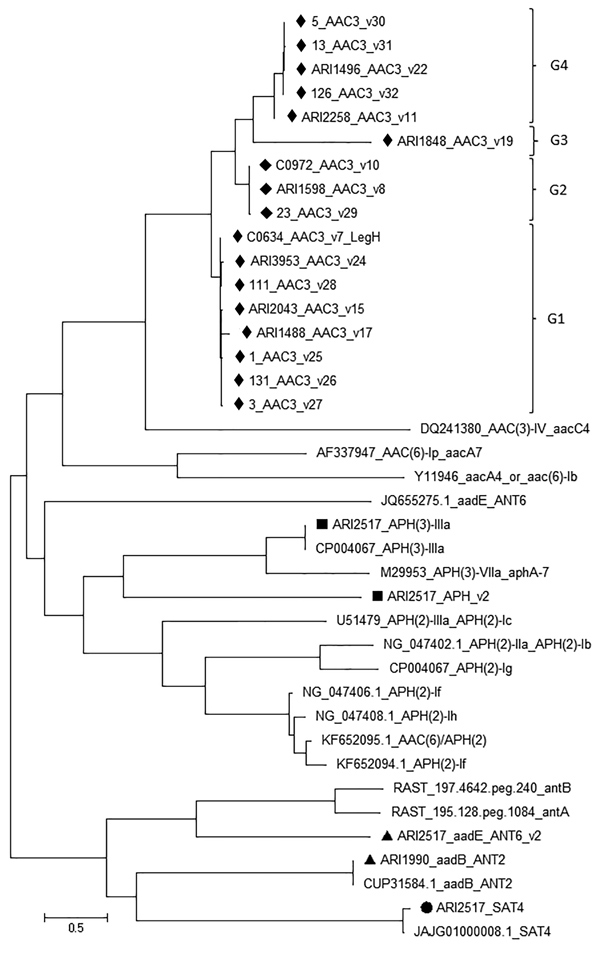
Figure 4. Maximum-likelihood tree of all aminoglycoside resistance determinants found in Campylobacter jejuni CC464, CC353, CC354, and CC574 isolates, Scotland. The tree was built using MEGA7 (https://www.megasoftware.net). The variants (v) listed in G1, G2, G3, G4 groups commonly occur in the 4 CCs. Scale bar indicates nucleotide substitutions per site. Aminoglycoside resistance gene variants identified in representative isolates of 4 CCs are shown by different symbols: black diamonds, aac3; black squares, aph; black triangles, ant; black circle, sat. CC, clonal complex.
Page created: June 17, 2019
Page updated: June 17, 2019
Page reviewed: June 17, 2019
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.