Volume 27, Number 10—October 2021
Dispatch
Confirmation of Rickettsia conorii Subspecies indica Infection by Next-Generation Sequencing, Shandong, China
Figure 2
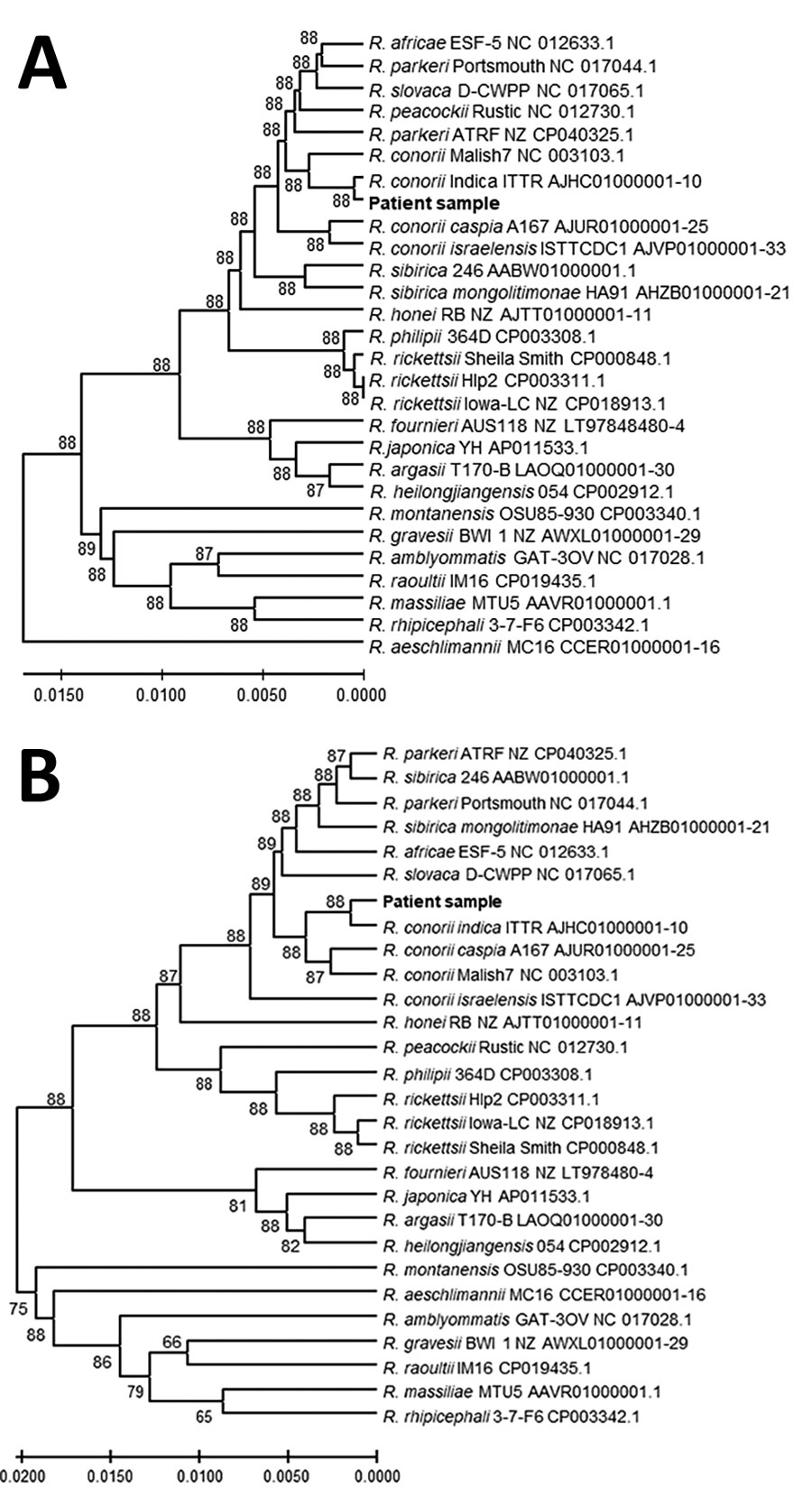
Figure 2. Genetic relationships of the spotted fever group rickettsia detected in blood of patient 1 in study of confirmation of Rickettsia conorii subspecies indica infection by next-generation sequencing, Shandong, China. This analysis used concatenated sequences from 27 spotted fever rickettsial genomes homologous to the patient sequences (shown in bold text). A) Analysis of 1,379 positions in the tRNA-associated sequences; B) analysis of 1,519 positions in the protein gene–associated sequences. Each tree was constructed upon concatenation of 6 different genome sites (Appendix Table 2); the consensus of reads from sites with overlapping reads was used. The evolutionary relationships were inferred by using UPGMA implemented in MEGA X (15). The optimal trees are shown. The percentage of replicate trees in which the taxa clustered together in the bootstrap test (500 replicates) are shown next to the branches. The evolutionary distances computed by using the Kimura 2-parameter method are in the units of the number of base substitutions per site. The proportion of sites where >1 unambiguous base is present in >1 sequence for each descendent clade is shown next each internal node in the tree. All ambiguous positions were removed for each sequence pair (pairwise deletion option). Scale bars indicate the percentage of nucleotide variation between the sequences.
References
- Chen R, Kou Z, Xu L, Cao J, Liu Z, Wen X, et al. Analysis of epidemiological characteristics of four natural-focal diseases in Shandong Province, China in 2009-2017: A descriptive analysis. PLoS One. 2019;14:
e0221677 . DOIPubMedGoogle Scholar - Qin XR, Han HJ, Han FJ, Zhao FM, Zhang ZT, Xue ZF, et al. Rickettsia japonica and novel Rickettsia species in ticks, China. Emerg Infect Dis. 2019;25:992–5. DOIPubMedGoogle Scholar
- Zhang R, Zhao A, Wang X, Zhang Z. Diversity of tick species on domestic animals in Shandong Province, China, using DNA barcoding. Exp Appl Acarol. 2017;73:79–89. DOIPubMedGoogle Scholar
- Fang LQ, Liu K, Li XL, Liang S, Yang Y, Yao HW, et al. Emerging tick-borne infections in mainland China: an increasing public health threat. Lancet Infect Dis. 2015;15:1467–79. DOIPubMedGoogle Scholar
- Schlaberg R, Chiu CY, Miller S, Procop GW, Weinstock G; Professional Practice Committee and Committee on Laboratory Practices of the American Society for Microbiology; Microbiology Resource Committee of the College of American Pathologists. Microbiology Resource Committee of the College of American Pathologists. Validation of metagenomic next-generation sequencing tests for universal pathogen detection. Arch Pathol Lab Med. 2017;141:776–86. DOIPubMedGoogle Scholar
- Zhu Y, Fournier PE, Eremeeva M, Raoult D. Proposal to create subspecies of Rickettsia conorii based on multi-locus sequence typing and an emended description of Rickettsia conorii. BMC Microbiol. 2005;5:11. DOIPubMedGoogle Scholar
- Biswal M, Zaman K, Suri V, Gopi S, Kumar A, Gopi T, et al. Molecular confirmation & characterization of Rickettsia conorii in north India: A report of three cases. Indian J Med Res. 2020;151:59–64. DOIPubMedGoogle Scholar
- Torina A, Fernández de Mera IG, Alongi A, Mangold AJ, Blanda V, Scarlata F, et al. Rickettsia conorii Indian tick typhus strain and R. slovaca in humans, Sicily. Emerg Infect Dis. 2012;18:1008–10. DOIPubMedGoogle Scholar
- Ghafar A, Khan A, Cabezas-Cruz A, Gauci CG, Niaz S, Ayaz S, et al. An assessment of the molecular diversity of ticks and tick-borne microorganisms of small ruminants in Pakistan. Microorganisms. 2020;8:
E1428 . DOIPubMedGoogle Scholar - Phongmany S, Rolain JM, Phetsouvanh R, Blacksell SD, Soukkhaseum V, Rasachack B, et al. Rickettsial infections and fever, Vientiane, Laos. Emerg Infect Dis. 2006;12:256–62. DOIPubMedGoogle Scholar
- Guo LP, Jiang SH, Liu D, Wang SW, Chen CF, Wang YZ. Emerging spotted fever group rickettsiae in ticks, northwestern China. Ticks Tick Borne Dis. 2016;7:1146–50. DOIPubMedGoogle Scholar
- Song S, Chen C, Yang M, Zhao S, Wang B, Hornok S, et al. Diversity of Rickettsia species in border regions of northwestern China. Parasit Vectors. 2018;11:634. DOIPubMedGoogle Scholar
- Hazihan W, Dong Z, Guo L, Rizabek K, Askar D, Gulzhan K, et al. Molecular detection of spotted fever group rickettsiae in ticks parasitizing pet dogs in Shihezi City, northwestern China. Exp Appl Acarol. 2019;77:73–81. DOIPubMedGoogle Scholar
- Zhang G, Zheng D, Tian Y, Li S. A dataset of distribution and diversity of ticks in China. Sci Data. 2019;6:105. DOIPubMedGoogle Scholar
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol. 2018;35:1547–9. DOIPubMedGoogle Scholar
1These authors contributed equally to this article.