Volume 27, Number 8—August 2021
Research Letter
Linezolid- and Multidrug-Resistant Enterococci in Raw Commercial Dog Food, Europe, 2019–2020
Figure
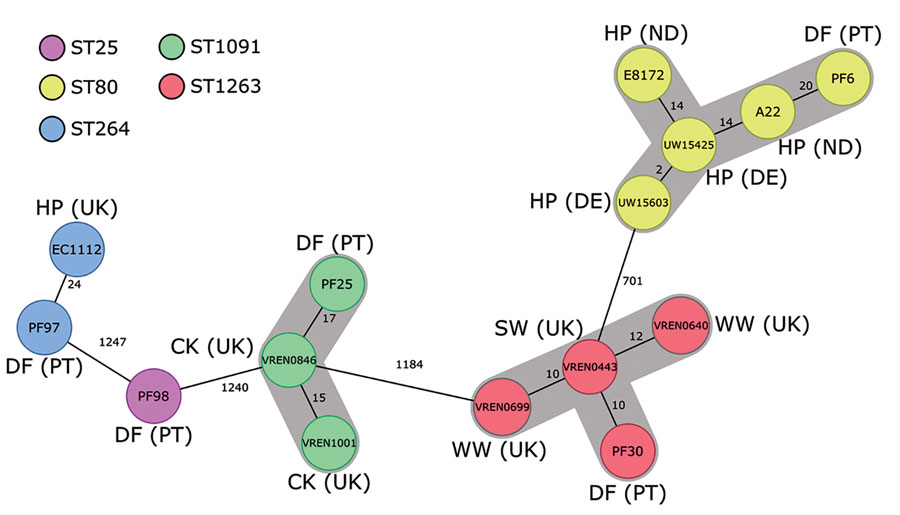
Figure. Minimum-spanning tree based on the core-genome multilocus sequence typing (cgMLST) data from Enterococcus faecium isolates (n = 15) from different sources in Europe. The tree is based on cgMLST (1,423 genes) analyses made with Ridom SeqSphere+ version 7.2 software (https://www.ridom.de/seqsphere). Each circle represents 1 allele profile. The numbers on the connecting lines represent the number of cgMLST allelic differences between 2 isolates. Sequence types are shown in colored circles (see key); numbers in circles are isolate identifications. Gray shading around nodes indicates clusters of closely related isolates (<20). CK, chicken; DE, Denmark; DF, dog food; HP, hospitalized patient; PT, Portugal; ST, sequence type; SW, swine; UK, United Kingdom; WW, wastewater.
1These authors were co–principal investigators.
2These authors are active EFWISG members.