Volume 28, Number 12—December 2022
Dispatch
Highly Diverse Arenaviruses in Neotropical Bats, Brazil
Figure 2
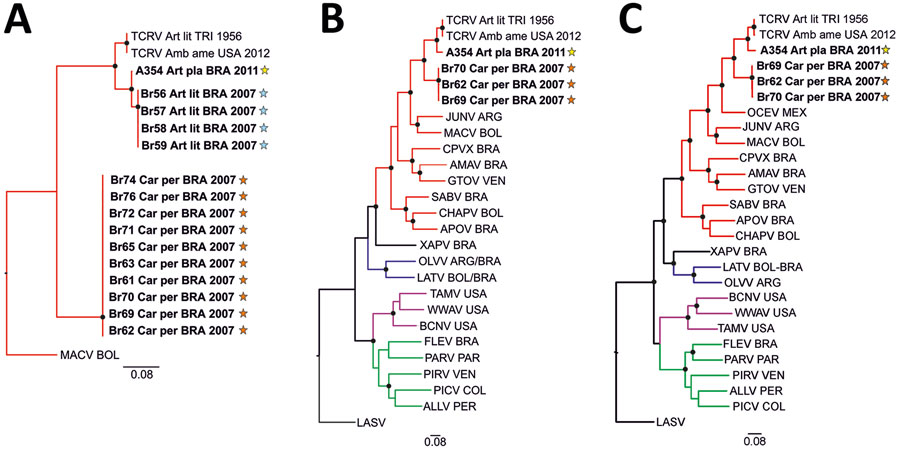
Figure 2. Phylogenetic analyses of highly diverse arenaviruses in neotropical bats, Brazil. Maximum-likelihood consensus trees compare partial RNA-dependent RNA polymerase genes (A), complete large (L) segment genes (B), and complete small (S) segment genes (C) from arenaviruses detected in Artibeus and Carollia spp. bats. Phylogenetic trees were generated using MEGA X software (https://www.megasoftware.net). Bold indicates sequences obtained from this study. Stars indicate regions where arenavirus-positive bat hosts were detected (Figure 1, panel B). Black dots at tree nodes represent bootstrap values >75% (1,000 replicates). Green lines indicate clade A new world arenaviruses, red lines indicate clade B new world arenaviruses, blue lines indicate clade C new world arenaviruses, and purple lines indicate recombinant new world arenaviruses (tentative clade D) (14). GenBank sequences used for comparisons and virus abbreviations (accession nos.): TCRV_Art_lit_TRI_1956, Tacaribe virus strain TRVL-11573 L segment (MT081317) and S segment (MT081316.1); TCRV_Amb_ame_USA_2012, Tacaribe virus Florida L segment (KF923401) and S segment (KF923400); OCEV, Ocozocoautla de Espinosa Virus S segment (JN897398; L segment not available); JUNV, Junin virus L segment (AY35802) and S segment (NC_005081.1); MACV, Machupo virus L segment (AY358021) and S segment (NC_005078.1); CPVX, Cupixi virus L segment (AY216519) and S segment (NC_010254.1); AMAV, Amapari virus L segment (AY216517) and S segment (NC_010247.1); GTOV, Guanarito virus L segment (AY358024) and S segment (NC_005077.1); SABV, Sabia virus L segment (AY358026) and S segment (NC_006317.1); APOV, Apore virus L segment (MF317491) and S segment (MF317490); CHAPV, Chapare virus L segment (EU260464) and S segment (EU260463.1); XAPV, Xapuri virus L segment (MG976577) and S segment (NC_055439.1), LATV, Latino virus L segment (EU627612) and S segment (NC_010758.1); OLVV, Oliveros virus L segment (AY216514) and S segment (NC_010248.1); BCNV, Bear Canyon virus L segment (AY924390) and S segment (NC_010256.1); WWAV, Whitewater Arroyo virus L segment (AY924395) and S segment (NC_010700.1); TAMV, Tamiami virus L segment (AY924393) and S segment (NC_010701.1); FLEV, Flexal virus L segment (EU627611) and S segment (NC_010757.1); PARV, Parana virus L segment (EU627613) and S segment (NC_010756.1); PIRV, Pirital virus L segment (AY494081) and S segment (NC_005894.1); PICV, Pichinde virus L segment (AF427517) and S segment (MK896487.1); LASV, Lassa virus L segment (U73034) and S segment (NC_004296.1); ALLV, Allpahuayo virus L segment (AY216502) and S segment (NC_010253.1). Origins of arenaviruses are indicated for each sample: ARG, Argentina; BOL, Bolivia; BRA, Brazil; COL, Colombia; PER, Peru; TRI, Trinidad; USA, United States of America; VEN, Venezuela. Scale bars indicate nucleotide substitutions per site.
References
- Guth S, Mollentze N, Renault K, Streicker DG, Visher E, Boots M, et al. Bats host the most virulent-but not the most dangerous-zoonotic viruses. Proc Natl Acad Sci U S A. 2022;119:
e2113628119 . DOIPubMedGoogle Scholar - Charrel RN, de Lamballerie X. Zoonotic aspects of arenavirus infections. Vet Microbiol. 2010;140:213–20. DOIPubMedGoogle Scholar
- Downs WG, Anderson CR, Spence L, Aitken THG, Greenhall AH. Tacaribe virus, a new agent isolated from Artibeus bats and mosquitoes in Trinidad, West Indies. Am J Trop Med Hyg. 1963;12:640–6. DOIPubMedGoogle Scholar
- Malmlov A, Seetahal J, Carrington C, Ramkisson V, Foster J, Miazgowicz KL, et al. Serological evidence of arenavirus circulation among fruit bats in Trinidad. PLoS One. 2017;12:
e0185308 . DOIPubMedGoogle Scholar - Cogswell-Hawkinson A, Bowen R, James S, Gardiner D, Calisher CH, Adams R, et al. Tacaribe virus causes fatal infection of an ostensible reservoir host, the Jamaican fruit bat. J Virol. 2012;86:5791–9. DOIPubMedGoogle Scholar
- Sayler KA, Barbet AF, Chamberlain C, Clapp WL, Alleman R, Loeb JC, et al. Isolation of Tacaribe virus, a Caribbean arenavirus, from host-seeking Amblyomma americanum ticks in Florida. PLoS One. 2014;9:
e115769 . DOIPubMedGoogle Scholar - Moreno H, Möller R, Fedeli C, Gerold G, Kunz S. Comparison of the innate immune responses to pathogenic and nonpathogenic clade B New World arenaviruses. J Virol. 2019;93:e00148–19. DOIPubMedGoogle Scholar
- Holzerland J, Leske A, Fénéant L, Garcin D, Kolakofsky D, Groseth A. Complete genome sequence of Tacaribe virus. Arch Virol. 2020;165:1899–903. DOIPubMedGoogle Scholar
- Vieth S, Drosten C, Lenz O, Vincent M, Omilabu S, Hass M, et al. RT-PCR assay for detection of Lassa virus and related Old World arenaviruses targeting the L gene. Trans R Soc Trop Med Hyg. 2007;101:1253–64. DOIPubMedGoogle Scholar
- Grande-Pérez A, Martin V, Moreno H, de la Torre JC. Arenavirus quasispecies and their biological implications. Curr Top Microbiol Immunol. 2016;392:231–76. DOIPubMedGoogle Scholar
- Hoffmann C, Wurr S, Pallasch E, Bockholt S, Rieger T, Günther S, et al. Experimental Morogoro virus infection in its natural host, Mastomys natalensis. Viruses. 2021;13:851. DOIPubMedGoogle Scholar
- Jones ML. Longevity of captive mammals. Zool Garten N. F. Jena. 1982;52;113–28 [cited 2022 Nov 10]. http://www.rhinoresourcecenter.com/pdf_files/125/1256468598.pdf
- Cajimat MNB, Milazzo ML, Bradley RD, Fulhorst CF. Ocozocoautla de espinosa virus and hemorrhagic fever, Mexico. Emerg Infect Dis. 2012;18:401–5. DOIPubMedGoogle Scholar
- Radoshitzky SR, Bào Y, Buchmeier MJ, Charrel RN, Clawson AN, Clegg CS, et al. Past, present, and future of arenavirus taxonomy. Arch Virol. 2015;160:1851–74. DOIPubMedGoogle Scholar