Volume 30, Number 10—October 2024
Research
Bartonella spp. in Phlebotominae Sand Flies, Brazil
Figure 2
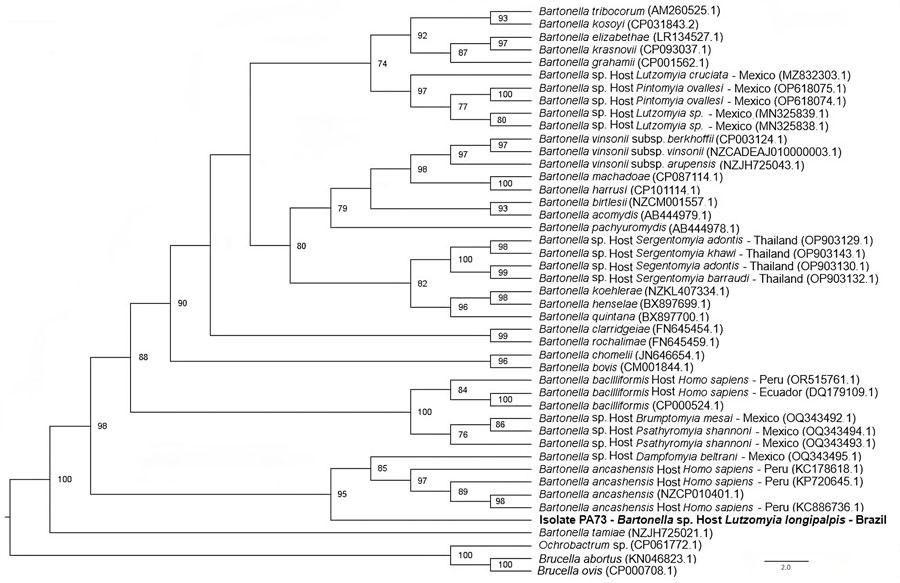
Figure 2. Phylogenetic tree based on an alignment of 380 bp-length of the gltA sequences obtained from phlebotomine sand flies collected in Brazil (bold) and reference sequences. Tree was created using the maximum-likelihood method and generalized time reversible plus invariate sites plus gamma as the evolutionary model. Ochrobactrum sp., Brucella ovis, and Brucella abortus were used as outgroups. Only bootstrap values >70 are shown. GenBank accession numbers are provided in parentheses.
Page created: September 18, 2024
Page updated: September 23, 2024
Page reviewed: September 23, 2024
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.