Volume 30, Number 10—October 2024
Research Letter
SARS-CoV-2 and Other Coronaviruses in Rats, Berlin, Germany, 2023
Figure 2
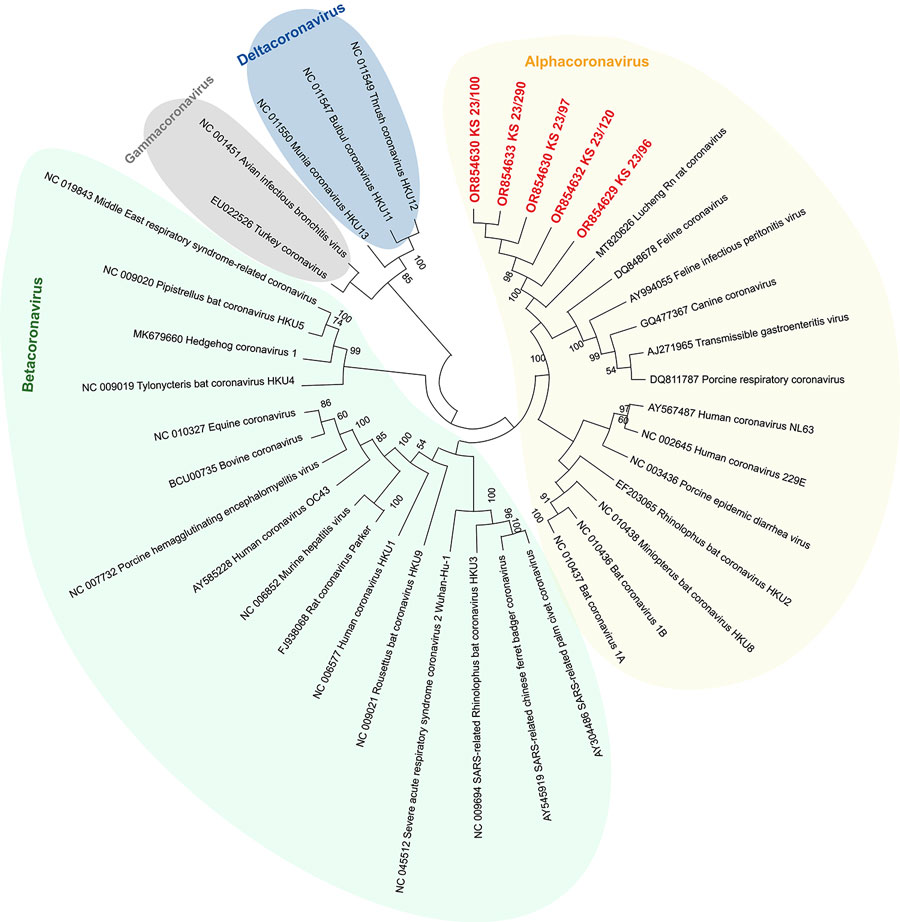
Figure 2. Phylogenetic analysis of SARS-CoV-2 and other coronaviruses in rats, Berlin, Germany, 2023. Partial sequences of the RNA-dependent RNA polymerase gene from coronaviruses isolated from rats in Berlin (red text) were compared with other coronavirus sequences obtained from GenBank. Background colors indicate viruses belonging to the same coronavirus genus. The maximum-likelihood tree was calculated by using MEGA X software (https://www.megasoftware.net). Statistical support for nodes was obtained by bootstrapping (1,000 replicates); only bootstrap values >50% are shown. GenBank accession numbers are provided. Tree not drawn to scale.
Page created: August 13, 2024
Page updated: September 24, 2024
Page reviewed: September 24, 2024
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.