Volume 30, Number 8—August 2024
Research
Geographic Distribution of Rabies Virus and Genomic Sequence Alignment of Wild and Vaccine Strains, Kenya
Abstract
Rabies, a viral disease that causes lethal encephalitis, kills ≈59,000 persons worldwide annually, despite availability of effective countermeasures. Rabies is endemic in Kenya and is mainly transmitted to humans through bites from rabid domestic dogs. We analyzed 164 brain stems collected from rabid animals in western and eastern Kenya and evaluated the phylogenetic relationships of rabies virus (RABV) from the 2 regions. We also analyzed RABV genomes for potential amino acid changes in the vaccine antigenic sites of nucleoprotein and glycoprotein compared with RABV vaccine strains commonly used in Kenya. We found that RABV genomes from eastern Kenya overwhelmingly clustered with the Africa-1b subclade and RABV from western Kenya clustered with Africa-1a. We noted minimal amino acid variances between the wild and vaccine virus strains. These data confirm minimal viral migration between the 2 regions and that rabies endemicity is the result of limited vaccine coverage rather than limited efficacy.
Rabies, a viral disease that causes encephalitis, is consistently deadly in exposed humans who are not vaccinated or promptly treated with postexposure prophylaxis (PEP). Rabies was recognized in Egypt around 2300 BCE (1), and a written account of the disease was included in the Laws of Eshnunna in Mesopotamia (2). From that region, rabies spread to Europe and then to Africa, following patterns of human colonization (3). In 1768, rabies was described in the Americas, occurring as an epizootic in Boston (E.C. Ramsey, honors thesis, Macalester College, 2017, https://digitalcommons.macalester.edu/intlstudies_honors/28).
Rabies virus (RABV) is transmitted through the bite of an infected animal, which inoculates the virus at the bite site. The virus travels from the inoculation site to the central nervous system (CNS), where it multiplies and migrates to the salivary glands, after which another animal or human can be inoculated through a bite. RABV modifies the behavior of its host, which becomes exceptionally aggressive, thus increasing the odds of conflict and biting (4). Once clinical signs appear in an infected host, fatality is nearly always certain (5). The primary mechanism of lethality involves direct targeting of the CNS via mechanisms that remain poorly defined (6).
RABV belongs to the genus Lyssavirus, which also includes Lagos bat virus, Mokola virus, Duvenhage virus, European bat virus 1 and 2, and Australian bat lyssavirus, among others (7). The primary viral reservoirs are members of the orders Carnivora (e.g., dogs) and Chiroptera (bats), whereas humans are dead-end hosts (8). Compared with other lyssaviruses, RABV is by far the most commonly reported (9), causing ≈59,000 human deaths annually (10), >99% of which are associated with dog-mediated transmissions (9). In rare human cases, rabies has been transmitted through non–bite-associated processes, such as organ transplants and laboratory exposures (11,12).
Rabies has 2 major epidemiologic cycles: the urban cycle, in which dogs are the major reservoirs, and the sylvatic cycle, in which wild animals are the primary reservoirs (13). The urban cycle predominates in Africa, Asia, and Central and South America (13). The United States and some countries in Europe have used oral rabies vaccine programs to substantially reduce the sylvatic cycle (14). Nonetheless, rabies remains a global public health and veterinary challenge, particularly in developing countries (15).
RABV can be categorized into 2 major phylogenetic groups: bat-related and dog-related. Bat-related RABV is confined to the New World, but dog-related RABV is globally distributed (16–18). The dog-related group segregates into 6 distinct clades, which are designated as Africa-2, Asian, Africa-3, Arctic-related, Cosmopolitan, and Indian subcontinent (19). Three of those clades, Cosmopolitan (Africa-1 and Africa-4 subclades), Africa-2, and Africa-3, are found in Africa (2,20). The Cosmopolitan clade has >22 subclades that are widely distributed in >100 countries (9). The grouping of those clades is greatly affected by physical barriers that restrict gene flow (21). The Africa-1a subclade dominates in northern and eastern Africa, and Africa-1b predominates in central, eastern, and southern Africa. Africa-1c circulates in Madagascar, and Africa-4 was recently discovered in northern Africa (22). The Africa-2 clade predominantly circulates in West Africa, but the Africa-3 clade is sustained through a sylvatic cycle in South Africa (19,22).
Kenya has a >100-year history of rabies; the first case was reported in 1912 (23). Despite successful control efforts in the past, the disease has remained endemic because of challenges in sustaining vaccination programs (24,25). The number of human deaths attributable to rabies in Kenya is unknown (23), but some studies report estimates of >2,000 per year and others as low as 523 (26,27); the highest incidences have been reported in western and eastern Kenya (23). The main source of human rabies is the domestic dog (28). However, those human deaths are unjustifiable considering the availability of effective mass dog vaccination and therapeutic PEP for persons bitten by rabid animals (25,29). Kenya has an ambitious plan to end human rabies deaths by 2030; that plan combines mass dog vaccination and prompt provision of PEP to affected humans (26). Unfortunately, only 5% of health facilities have PEP, and mass dog vaccination has fallen short of the target (25,26).
RABV, a negative-stranded RNA virus with a genome size of ≈12 kilobases, encodes 5 proteins: nucleoprotein (N), matrix protein (M), phosphoprotein (P), glycoprotein (G), and large (L) protein or polymerase (30,31). The N, P, and L proteins comprise the ribonucleoprotein (RNP) complex and the M and G proteins are involved in virus assembly and budding (32). The N gene is responsible for transcription and replication and shows antigenic variation that is used for strain discrimination (7,33,34–36). Since the 1880s, when Louis Pasteur developed a RABV vaccine (37), vaccination has played a crucial role in controlling rabies through production of neutralizing antibodies against the RNP and G antigens (38,39). G and N are crucial components for rabies vaccine effectiveness (38,39), and monitoring antigenic variations between vaccine strains and the G and N genes of circulating wild strains can help determine whether the existing vaccine strains remain effective (33,40). We assessed genetic diversity of wild RABV strains collected from 2 rabies hotspots in Kenya to inform current and future vaccination and PEP efforts.
Study Areas and Sample Cohort
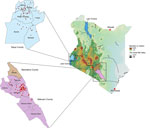
Veterinary technicians collected postmortem brain stems from suspected rabid animals in Kenya’s Siaya County, in the western region, and Makueni County, in the eastern region (Figure 1). Sample collection was approved by the Kenya Medical Research Institute’s Scientific and Ethical Research Unit under a rabies surveillance protocol (no. KEMRI/SERU/CGHR 046/3268).
We used Anigen Rapid Rabies Ag Kits (BioNote Inc., https://www.bionote.co.kr) to confirm RABV at the collection sites. That assay has a sensitivity of 92% (95% CI 85.9%–95.6%) and specificity of 100% (95% CI 93.4%–100%) compared with the reference standard fluorescent antibody test and is recommended for confirming clinical rabies cases in animals (41). We collected a total of 164 RABV-positive brain stems. We transported brainstems to the laboratory in liquid nitrogen and stored at −80°C until we performed nucleic acid purification.
Nucleic Acid Isolation
We used the Bullet Blender (Next Advance, https://www.nextadvance.com) to homogenize ≈20 mg brainstem with 3-mm ceramic beads in TRIzol reagent (Thermo Fisher Scientific, https://www.thermofisher.com). We cooled samples at room temperature before centrifuging at 12,000 × g for 3 minutes at 4°C. We used 200 μL of the aqueous phase to extract nucleic acids by using MagMAX CORE Purification Kit (ThermoFisher Scientific), then eluted extracted nucleic acids in a total volume of 60 µL.
Molecular Detection of RABV by qRT-PCR
We used quantitative reverse transcription PCR (qRT-PCR) to screen nucleic acids for RABV by targeting the large (L) gene using forward primer 5′-GGTTTCCGGDGCYGTDCCTC-3′, reverse primer 5′-CCTAGGGGAGACYTTGCCRT-3′ primer, and a 6FAM-CCCGTCAYATAGGGTCRGCTCARGGGC‐BBQ probe. The qRT-PCR reaction comprised 4 μL of the nucleic acid, 10 μL of 2x SensiFAST Master mix (Meridian Bioscience, https://www.meridianbioscience.com), 0.8 μL forward and reverse primer mix at a concentration of 10 μM each, 0.4 μL of TaqMan QSY probe (Applied Biosystems/Thermo Fisher Scientific) at a concentration of 10 μM, 3.4 μL of nuclease-free water, 0.4 μL RiboSafe RNase inhibitor (Thermo Fisher Scientific), and 0.2 μL reverse transcriptase. We used an ABI 7500 (Applied Biosystems) for amplification using a reverse transcription cycle at 45°C for 10 minutes and 95°C for 3 minutes to inactivate the reverse transcriptase, followed by 40 cycles of 95°C for 30 seconds and 60°C for 1 minute. Each reaction included a positive control of a known RABV-positive sample and PCR water as a nontemplate negative control.
Whole-Genome Sequencing
We used the TURBO DNase kit (ThermoFisher Scientific) to deplete extracted nucleic acids of host genomic DNA. We used sequence-independent, single-primer amplification to amplify viral RNA as previously described (42), with subsequent modifications (43). We then reversed transcribed the first cDNA strand by using the LunaScript RT SuperMix kit (New England Biolabs, https://www.neb.com) and the JH17N8 primer (5′-GTTTCCCAGTAGGTCTCNNNNNNNN-3′), which contained a degenerate 8-mer sequence at the 3′ end. We generated the second cDNA strand by amplification using NEB Next Ultra II Q5 master mix (New England Biolabs) with 10 μM of a specific primer JHP21 (5′-GTTTCCAGTAGGTCTC-3′) and the following thermal cycling regimen: 1 cycle of 94°C for 3 minutes, 25°C for 30 seconds, 72°C for 1 minute, followed by 35 cycles of 94°C for 30 seconds, 55°C for 30 seconds, and 72°C for 1 minute. We included a final extension step of 72°C for 1 minute, followed by cooling to 4°C. We purified the synthesized dsDNA by using Agencourt AMPure XP beads (Beckman Coulter, https://www.beckmancoulter.com), which we then used to prepare sequence libraries using the Colibri ES DNA Library preparation kit (ThermoFisher Scientific). We quantified libraries by using a Qubit dsDNA HS Assay kit (Invitrogen/Thermo Fisher Scientific) and measured the average library size on the 4200 TapeStation System (Agilent Technologies, https://www.agilent.com). We adjusted the pooled library to 4 nM concentration and then denatured with 0.2 normal sodium hydroxide and further diluted to 9.5 pM. We spiked 5% of PhiX v3 Control Library (Illumina, https://www.illumina.com) into the pool and sequenced the library in pairs using 600 cycles v3 on the Miseq or P3 reagents on the NextSeq 2000 platform (Illumina).
Genome Assembly, Clade Classification, and Phylogenetic Reconstruction
We assessed the quality of the raw sequences in FastQC v0.12.1 (https://github.com/s-andrews/FastQC/releases) and then processed by using the ngs_mapper pipeline (https://github.com/VDBWRAIR/ngs_mapper), which removes low-quality reads (<Q20), failed reads, sequencing adapters, and short reads (<50 nt) by using trimmomatic v0.35 (https://github.com/usadellab/Trimmomatic/releases) and cutadapt v1.9.1 (https://gensoft.pasteur.fr/docs/cutadapt/1.9.1/index.html). We then mapped the filtered reads against a RABV genome from Tanzania (GenBank accession no. KY210291) that had the closest homology to our sample sequences by using bwa v0.7.12 (https://github.com/lh3/bwa/releases). We used Samtools v0.1.19 (https://sourceforge.net/projects/samtools/files/samtools/0.1.19) to create pileups from the read alignments and the consensus genome and generated a variant call format file and coverage visualizations by using several Python scripts (Python Software Foundation, https://www.python.org) housed within the pipeline.
To determine whether the generated whole genomes add value to genomes generated from the N and G genes, we used the RABV-GLUE tool (http://rabv-glue.cvr.gla.ac.uk) to assign the RABV to major and minor clades. Further classification of the whole genomes into lineages was performed using MADDOG (44).
To establish the phylogenetic relationships between RABV from Kenya in the context of Africa, we obtained a comprehensive subset of curated, annotated, and published RABV datasets from Africa from the Bacterial and Viral Bioinformatics Resource Center (BV-BRC; https://www.bv-brc.org). We aligned the complete RABV polyprotein, the entire N protein, and the entire G protein of the study genomes and context samples in CLC Genomics workbench version 8.5.1 and the Muscle plugin (QIAGEN, https://www.qiagen.com). To avoid using sequences from recombinations, we ran the aligned sequences in GARD software (Datamonkey, https://www.datamonkey.org) by using the Hyphy package (Datamonkey). We performed phylogenetic inference by using the maximum-likelihood method in IQ-TREE version 2.2.0 (http://www.iqtree.org) and nucleotide substitution models built into ModelFinder in IQ-TREE. We evaluated node support with a combination of approximate likelihood tests and ultrafast bootstraps with 1,000 replicates, each computed in IQ-TREE (45). We visualized and annotated the resulting phylogenetic trees by using FigTree version 1.4.2 (http://tree.bio.ed.ac.uk/software/Figtree).
Analysis of Amino Acid Variation at RABV Vaccine Target Sites
To identify amino acid variations at the N and G vaccine target sites of the study genomes and the vaccine strains, we performed sequence alignment of the G (n = 142) and N (n = 144) proteins by using CLC Genomics Main Workbench (QIAGEN). We aligned study sample sequences to 3 commonly used RABV vaccine strains: Pitman-Moore (PM; GenBank accession no. DQ099525), Pasteur virus (PV; GenBank accession no. M13215), and challenge virus standard (CVS; accession no. AF406696 for N and AF406694 for G).
Study Sample Demographics
Domestic dogs contributed most (65%) study samples, followed by cows (18%), and goats (14%). Other species accounted for only 1% of samples (Appendix 1 Table 1)
RABV Geographic Restriction in Kenya
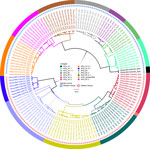
Of the 164 brainstems collected, 144 samples with genome lengths ranging from 10,024 to 11,923 nt were used for whole-genome analysis. From whole-genome sequences, we also extracted 142 N and 144 G genes for single-gene analysis. By RABV-GLUE, all genomes belonged to the Cosmopolitan clade, either Africa-1a (71/144) or Africa-1b (73/144) subclades. Further interrogation of the clades using the MAADOG lineage typing tool revealed 14 distinct lineages, 8 in western Kenya, 6 in eastern Kenya, including 1 unclassified lineage in eastern Kenya (Figure 2).
We accessed the BV-BRC database on November 18, 2022, and retrieved 378 RABV polyproteins, 1,500 N, and 130 G sequences that met our inclusion criteria. We used RABV sequences from the study strains and strains subsampled from Africa to construct whole-genome phylogenetic trees (Appendix 2 Figures 1–3) and individual G and N gene trees (Appendix 2 Figures 4, 5). We compared the phylogenetic trees constructed using a relatively new method of whole-genome sequencing (has only few sequences in literature) with those generated from the old method of individual G and N genes analysis that has been the basis of RABV phylogeography. Both methods identified only the Cosmopolitan clade in our study genomes (Appendix 2 Figures 2, 4, 5). In addition, western Kenya samples (97.2%) branched with the Cosmopolitan Africa-1a subclade, and eastern Kenya samples (95.7%) branched with Cosmopolitan Africa-1b subclade (Appendix 2 Figures 2, 4, 5). Only 2 western Kenya genomes clustered with the Africa-1b subclade (Appendix 2 Figure 1) and only 3 eastern Kenya genomes clustered with the Africa-1a subclade (Appendix 2 Figure 3).
Within the Africa-1b subclade, the eastern Kenya samples appeared to have 2 distinct clusters, 1 major (n = 63) and 1 minor (n = 8) (Appendix 2 Figure 1). In both clusters, the study samples branched with Tanzania genomes. The western Kenya genomes (n = 69) were more homogeneous (Appendix 2 Figure 3). We deposited raw sequence data from this study in the National Center for Biotechnology Information (NCBI; https://www.ncbi.nlm.nih.gov) Sequence Read Archive (BioProject nos. OR256801 and OR270967–1061).
Comparison of Circulating Wild RABV and Vaccine Strains
Compared with sequences of the commonly used RABV vaccine strains in Kenya (PM, PV, and CVS), we noted no amino acid substitutions at the antigenic sites of the G gene, located between amino acids 20–439 (Appendix 1 Table 2). However, sequence homology outside the antigenic sites varied from 92.2% for PM to 93.3% for CVS and 93.0% for PV. In contrast, 2 study samples, Kenya/SIA-CAN-018/2021 and Kenya/SIA-BOV-007/202, were variant at antigenic site II of the N gene, in which alanine was substituted by valine at position 315 (A315V), but the 3 vaccine strains and all the other study samples had alanine in that position. In addition, although all the study samples, PM, and CVS had valine at position 379 of antigenic site III, PV was variant with alanine (V379A). Outside the antigenic sites of the N gene, sequence homology ranged from 98.45% to 99.11% for PM, 97.78% to 98.67% for CVS, and 97.56% to 98.23% for PV.
In this study, we used samples collected from 2 geographically isolated counties, Siaya in western Kenya and Makueni in eastern Kenya. We chose those 2 counties because they continuously report the highest rabies incidences in humans (23). Of the 164 brainstem study samples, 107 were from domestic dogs (Appendix 1 Table 1). Moreover, the other 57 samples came from animals with a history of having been bitten by a rabid dog, underscoring the role domestic dogs play in rabies transmission.
Previous phylogenetic studies, performed using single G and N genes, have indicated that RABV in Africa falls into several regional groups and that viruses from eastern Africa are genetically distinct from those in the western, central, and southern parts of the continent (21). Before this study, NCBI included 43 RABV sequences from Kenya (46–48). In a previous study (46), N and G genes from RABV samples obtained during an outbreak of rabies in African wild dogs in Kenya and Tanzania appeared to be identical. The authors concluded that the outbreak was most likely caused by a viral variant frequently found in domestic dogs in Kenya and Tanzania. A subsequent study that used N and G gene sequences reported existence of 2 Cosmopolitan subclades (47), namely Africa-1a and Africa-1b. Subsequent research revealed predominance of Africa-1a in western Kenya, but Africa-1b was more commonly observed in the eastern part of the country (48).
We added 144 whole-genome sequences from this study to the NCBI Sequence Read Archive. We used those genomes to further characterize the diversity of RABV from eastern and western Kenya. We also determined whether available rabies vaccines would confer protection. Our study confirms the inferences of previous research that shows an apparent geographic isolation between the RABV strains in eastern and western Kenya (48). Of the 3 major clades of RABV found in Africa, only the Cosmopolitan clade was detected by whole genomes (Appendix 2 Figures 1–3) and individual N and G genes (Appendix 2 Figures 4, 5). Predominantly, RABV from eastern Kenya clustered with the Africa-1b subclade, whereas RABV from western Kenya clustered with the Africa-1a subclade. That geographic isolation is probably because of the multiple landscape features that would restrict free movement of animals between the regions (Figure 1), thus promoting localized viral evolution. Similar geographic restriction has also been observed in raccoon-mediated rabies in the eastern United States (49). Outlier Africa-1b genomes that were in western Kenya (n = 2), and Africa-1a genomes (n = 3) that were in eastern Kenya indicate that geographic isolation is not absolute in the country (Appendix 2 Figures 1–3).
We found that RABV strains in western Kenya were closely related to each other and dissimilar from other Africa-1a subclade members in neighboring countries (Appendix 2 Figures 1–3). For example, the closest genome that was clearly distinct from the Kenya 1a subclade was from Sudan. Those observations suggest that the members of the Africa-1a circulating in Kenya have evolved separately, most likely from northern, central, and western regions of Africa. The other likely explanation is undersampling given that our study samples were collected in 2021 and 2022, but the other Africa-1a subgenomes were collected during 1986–2015. The Africa-1b genomes from Kenya were less homogeneous and branched into 2 groups: a major group of 63 genomes that clustered together and a minor group of 8 genomes that clustered with genomes that had been collected previously from Kericho and Nakuru, Kenya. However, using the MADDOG lineage typing tool, the western Kenya 1a subclade that was apparently homogeneous revealed 8 lineages, and the eastern Kenya subclade 1b segregated into 5 lineages (Figure 2), indicating that RABV accumulates mutations through the course of transmission. The finding of Africa-1b in Nakuru and Kericho, locales that are close to western Kenya, indicates an ongoing encroachment of the 1b subclade into western Kenya.
Several strains of RABV are used to manufacture vaccines (38). The choice of vaccine strain for use in a geographic region is informed by factors such as the circulating wild variants and species of animals being vaccinated (50). Live attenuated vaccines are given orally and are used in wild carnivores. Inactivated vaccines are used in humans and domestic animals, including dogs and cats. From the RABV genomes in our study, we evaluated whether the currently used RABV vaccines would confer protection (Appendix 1 Tables 2, 3). In Kenya, the available vaccines are derived from the PM, CVS, and PV RABV strains. An amino acid sequence alignment and comparison of the G sequences of the study genomes and the vaccine strains revealed 100% homology to the vaccine antigenic sites (Appendix 1 Table 2). A similar alignment and comparison using the N gene revealed a V315A replacement at antigenic site II in only 2 study samples. In addition, all the study samples and PM and CVS strains had valine at position 379 of antigenic site III, but the PV was variant with V379A (Appendix 1 Table 3). Although those replacements are too infrequent to effect vaccine efficacy, they raise concerns of potential cumulative changes that could eventually alter vaccine efficacy and underscore the need for continued monitoring of such changes.
In conclusion, we used whole-genome sequencing to define the genetic diversity of RABV in Kenya. Our data demonstrated the presence of localized viral lineages and limited viral migration between the 2 study regions. In addition, obtained data suggest that rabies endemicity is due to limited vaccine use because the sequences of the study strains do not greatly diverge from current vaccine strains. Moving forward, similar studies should expand to the other regions of Kenya to determine the generalizability of our findings. Nonetheless, the viral migration across the regions, though limited, reinforces the need for cross-county rabies surveillance systems in Kenya.
Ms. Wambugu is an MS student at the University of Embu. She has broad interests in whole-genome sequencing and bioinformatics.
Author contributions: E.W. performed all the assays, performed genome sequencing and associated bioinformatics analysis and interpretation, wrote the first draft, reviewed and edited the manuscript. K.G. assisted E.W. in sequencing, supervised the bioinformatics analysis and interpretation. S.K. assisted E.W. in data analysis and interpretation, reviewed and edited the manuscript. M.A.W. reviewed multiple drafts of the manuscripts, made suggestions in data presentation. C.M. was in charge of maintaining sample inventory, assisted E.W. in nucleic acids extraction and qRT-PCR quality assurance and quality control. L.M. and G.J. supervised field collection and shipping logistic to Kisumu Basic Science Laboratory. T.S.M. was the PI for the protocol that provided the rabies virus for the current study. He revised and edited the manuscript. B.C.S conceived the study and obtained funding that supported the study in Kenya. J.W. was Kenya’s principal investigator, supervised all aspects of the study, and worked with E.W. on the multiple versions of the manuscript.
Acknowledgments
We are grateful to the veterinary team that worked hard to trace rabid animals. We thank Rachel Githii for generating the map shown in Figure 1.
Research support was provided by Uniformed Services University grant HU00011920118 to B.C.S.
Material has been reviewed by the Walter Reed Army Institute of Research. There is no objection to its publication. The opinions or assertions contained herein are the private views of the author, and they are not to be construed as official, or as reflecting true views of the Department of the Army or the Department of Defense.
References
- Steele JH, Fernandez PJ. History of rabies and global aspects. In: Baer GM, editor. The natural history of rabies. Boca Raton (FL): CRC Press; 2017. p. 1–24.
- Dunlop RH, Williams DJ. Veterinary medicine: an illustrated history. St. Louis (MO): Mosby-Year Book, Inc.; 1996.
- Talbi C, Holmes EC, de Benedictis P, Faye O, Nakouné E, Gamatié D, et al. Evolutionary history and dynamics of dog rabies virus in western and central Africa. J Gen Virol. 2009;90:783–91. DOIPubMedGoogle Scholar
- Schnell MJ, McGettigan JP, Wirblich C, Papaneri A. The cell biology of rabies virus: using stealth to reach the brain. Nat Rev Microbiol. 2010;8:51–61. DOIPubMedGoogle Scholar
- Yousaf MZ, Qasim M, Zia S, Khan M, Ashfaq UA, Khan S. Rabies molecular virology, diagnosis, prevention and treatment. Virol J. 2012;9:50. DOIPubMedGoogle Scholar
- Fisher CR, Streicker DG, Schnell MJ. The spread and evolution of rabies virus: conquering new frontiers. Nat Rev Microbiol. 2018;16:241–55. DOIPubMedGoogle Scholar
- Walker PJ, Freitas-Astúa J, Bejerman N, Blasdell KR, Breyta R, Dietzgen RG, et al.; Ictv Report Consortium. ICTV virus taxonomy profile: Rhabdoviridae 2022. J Gen Virol. 2022;103:
001689 . DOIPubMedGoogle Scholar - Müller T, Freuling CM. Rabies in terrestrial animals. In: Fooks AR, Jackson AC, editors. Rabies: scientific basis of the disease and its management, 4th edition. London: Elsevier Academic Press; 2020. p. 195–230.
- Shipley R, Wright E, Lean FZX, Selden D, Horton DL, Fooks AR, et al. Assessing rabies vaccine protection against a novel lyssavirus, Kotalahti bat lyssavirus. Viruses. 2021;13:947. DOIPubMedGoogle Scholar
- World Health Organization. WHO expert consultation on rabies. WHO technical report series no. 931. Geneva: The Organization; 2005.
- Scott TP, Nel LH. Lyssaviruses and the fatal encephalitic disease rabies. Front Immunol. 2021;12:
786953 . DOIPubMedGoogle Scholar - Singh R, Singh KP, Cherian S, Saminathan M, Kapoor S, Manjunatha Reddy GB, et al. Rabies - epidemiology, pathogenesis, public health concerns and advances in diagnosis and control: a comprehensive review. Vet Q. 2017;37:212–51. DOIPubMedGoogle Scholar
- Meske M, Fanelli A, Rocha F, Awada L, Soto PC, Mapitse N, et al. Evolution of rabies in South America and inter-species dynamics (2009–2018). Trop Med Infect Dis. 2021;6:98. DOIPubMedGoogle Scholar
- Slate D, Algeo TP, Nelson KM, Chipman RB, Donovan D, Blanton JD, et al. Oral rabies vaccination in north america: opportunities, complexities, and challenges. PLoS Negl Trop Dis. 2009;3:
e549 . DOIPubMedGoogle Scholar - World Health Organization. WHO expert consultation on rabies. Second report. World Health Organ Tech Rep Ser. 2013;982:1–139.PubMedGoogle Scholar
- Zhou M, Zhou Z, Kia GSN, Gnanadurai CW, Leyson CM, Umoh JU, et al. Complete genome sequence of a street rabies virus isolated from a dog in Nigeria. Genome Announc. 2013;1:e00214–12. DOIPubMedGoogle Scholar
- Kuzmin IV, Shi M, Orciari LA, Yager PA, Velasco-Villa A, Kuzmina NA, et al. Molecular inferences suggest multiple host shifts of rabies viruses from bats to mesocarnivores in Arizona during 2001-2009. PLoS Pathog. 2012;8:
e1002786 . DOIPubMedGoogle Scholar - Caraballo DA, Lema C, Novaro L, Gury-Dohmen F, Russo S, Beltrán FJ, et al. A novel terrestrial rabies virus lineage occurring in South America: origin, diversification, and evidence of contact between wild and domestic cycles. Viruses. 2021;13:2484. DOIPubMedGoogle Scholar
- Troupin C, Dacheux L, Tanguy M, Sabeta C, Blanc H, Bouchier C, et al. Large-scale phylogenomic analysis reveals the complex evolutionary history of rabies virus in multiple carnivore hosts. PLoS Pathog. 2016;12:
e1006041 . DOIPubMedGoogle Scholar - Bourhy H, Reynes J-M, Dunham EJ, Dacheux L, Larrous F, Huong VTQ, et al. The origin and phylogeography of dog rabies virus. J Gen Virol. 2008;89:2673–81. DOIPubMedGoogle Scholar
- Brunker K, Hampson K, Horton DL, Biek R. Integrating the landscape epidemiology and genetics of RNA viruses: rabies in domestic dogs as a model. Parasitology. 2012;139:1899–913. DOIPubMedGoogle Scholar
- Sadeuh-Mba SA, Momo JB, Besong L, Loul S, Njouom R. Molecular characterization and phylogenetic relatedness of dog-derived Rabies Viruses circulating in Cameroon between 2010 and 2016. PLoS Negl Trop Dis. 2017;11:
e0006041 . DOIPubMedGoogle Scholar - Bitek AO, Osoro E, Munyua PM, Nanyingi M, Muthiani Y, Kiambi S, et al. A hundred years of rabies in Kenya and the strategy for eliminating dog-mediated rabies by 2030. AAS Open Res. 2019;1:23. DOIPubMedGoogle Scholar
- McDermott JJ, Kitala PM. The epidemiology of rabies in Machakos District, Kenya. Nairobi: University of Nairobi College of Agriculture and Veterinary Studies; 2003.
- Chuchu VM, Kitala PM, Bichanga P, Ksee D, Muturi M, Mwatondo A, et al. Rabies elimination in rural Kenya: need for improved availability of human vaccines, awareness and knowledge on rabies and its management among healthcare workers. Front Public Health. 2022;10:
769898 . DOIPubMedGoogle Scholar - Republic of Kenya, Ministry of Health and Ministry of Agriculture, Livestock and Fisheries. Strategic plan for the elimination of human rabies in Kenya 2014–2030. Nairobi: The Republic; 2014.
- Hampson K, Coudeville L, Lembo T, Sambo M, Kieffer A, Attlan M, et al.; Global Alliance for Rabies Control Partners for Rabies Prevention. Estimating the global burden of endemic canine rabies. PLoS Negl Trop Dis. 2015;9:
e0003709 . DOIPubMedGoogle Scholar - World Health Organization. WHO Expert Consultation on Rabies, WHO technical report series, no. 1012. Geneva: The Organization; 2018.
- World Health Organization. Rabies vaccines: WHO position paper, April 2018 - Recommendations. Vaccine. 2018;36:5500–3. DOIPubMedGoogle Scholar
- Luo Y, Zhang Y, Liu X, Yang Y, Yang X, Zhang D, et al. Complete genome sequence of a highly virulent rabies virus isolated from a rabid pig in south China. J Virol. 2012;86:12454–5. DOIPubMedGoogle Scholar
- Zhang G, Fu ZF. Complete genome sequence of a street rabies virus from Mexico. J Virol. 2012;86:10892–3. DOIPubMedGoogle Scholar
- Wunner WH, Conzelmann K-K. Rabies virus. In: Fooks AR, Jackson AC, editors. Rabies: scientific basis of the disease and its management, 4th edition. London: Elsevier Academic Press; 2020. p. 43–81.
- Wang W, Ma J, Nie J, Li J, Cao S, Wang L, et al. Antigenic variations of recent street rabies virus. Emerg Microbes Infect. 2019;8:1584–92. DOIPubMedGoogle Scholar
- Morimoto K, Hooper DC, Carbaugh H, Fu ZF, Koprowski H, Dietzschold B. Rabies virus quasispecies: implications for pathogenesis. Proc Natl Acad Sci U S A. 1998;95:3152–6. DOIPubMedGoogle Scholar
- de Almeida GL, Cargnelutti JF, Ries AS, Ferreira JC, Rosa JCA, Batista HBCR, et al. Sequence analysis of nucleoprotein gene reveals the co-circulation of lineages and sublineages of rabies virus in herbivorous in Rio Grande do Sul state, Brazil. Braz J Microbiol. 2020;51:837–46. DOIPubMedGoogle Scholar
- Zandi F, Goshadrou F, Meyfour A, Vaziri B. Rabies infection: an overview of lyssavirus–host protein interactions. Iran Biomed J. 2021;25:226–42. DOIPubMedGoogle Scholar
- Brightman C. Rabies: an acute viral infection. Trends Urol Men’s Heal. 2012;3:31–3. DOIGoogle Scholar
- Ajorloo M, Mirzaei H, Sadeghi Y, Tarban N, Soltani S, Mohammadi FS, et al. Evaluation and phylogenetic analysis of regular rabies virus vaccine strains. Arch Iran Med. 2018;21:101–10.PubMedGoogle Scholar
- Kim H-H, Yang D-K, Nah J-J, Song J-Y, Cho I-S. Comparison of the protective efficacy between single and combination of recombinant adenoviruses expressing complete and truncated glycoprotein, and nucleoprotein of the pathogenic street rabies virus in mice. Virol J. 2017;14:122. DOIPubMedGoogle Scholar
- Zhang G, Wang H, Mahmood F, Fu ZF. Rabies virus glycoprotein is an important determinant for the induction of innate immune responses and the pathogenic mechanisms. Vet Microbiol. 2013;162:601–13. DOIPubMedGoogle Scholar
- Tenzin T, Lhamo K, Rai PB, Tshering D, Jamtsho P, Namgyal J, et al. Evaluation of a rapid immunochromatographic test kit to the gold standard fluorescent antibody test for diagnosis of rabies in animals in Bhutan. BMC Vet Res. 2020;16:183. DOIPubMedGoogle Scholar
- Bohlander SK, Espinosa R III, Le Beau MM, Rowley JD, Díaz MO. A method for the rapid sequence-independent amplification of microdissected chromosomal material. Genomics. 1992;13:1322–4. DOIPubMedGoogle Scholar
- Wang D, Coscoy L, Zylberberg M, Avila PC, Boushey HA, Ganem D, et al. Microarray-based detection and genotyping of viral pathogens. Proc Natl Acad Sci U S A. 2002;99:15687–92. DOIPubMedGoogle Scholar
- Campbell K, Gifford RJ, Singer J, Hill V, O’Toole A, Rambaut A, et al. Making genomic surveillance deliver: A lineage classification and nomenclature system to inform rabies elimination. PLoS Pathog. 2022;18:
e1010023 . DOIPubMedGoogle Scholar - Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic ea. Mol Biol Evol. 2020;37:1530–4. DOIPubMedGoogle Scholar
- Kat PW, Alexander KA, Smith JS, Munson L. Rabies and African wild dogs in Kenya. Proc Biol Sci. 1995;262:229–33. DOIPubMedGoogle Scholar
- Brunker K, Jaswant G, Thumbi SM, Lushasi K, Lugelo A, Czupryna AM, et al. Rapid in-country sequencing of whole virus genomes to inform rabies elimination programmes. Wellcome Open Res. 2020;5:3. DOIPubMedGoogle Scholar
- Gigante CM, Yale G, Condori RE, Costa NC, Long NV, Minh PQ, et al. Portable rabies virus sequencing in canine rabies endemic countries using the Oxford Nanopore MinION. Viruses. 2020;12:1255. DOIPubMedGoogle Scholar
- Wheeler DC, Waller LA. Mountains, valleys, and rivers: The transmission of raccoon rabies over a heterogeneous landscape. J Agric Biol Environ Stat. 2008;13:388–406. DOIPubMedGoogle Scholar
- Metlin A, Paulin L, Suomalainen S, Neuvonen E, Rybakov S, Mikhalishin V, et al. Characterization of Russian rabies virus vaccine strain RV-97. Virus Res. 2008;132:242–7. DOIPubMedGoogle Scholar
Figures
Cite This ArticleOriginal Publication Date: July 15, 2024
Table of Contents – Volume 30, Number 8—August 2024
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
John N. Waitumbi, WRAIR-Africa, Kenya Medical Research Institute, Basic Science Laboratory, PO Box 54, Kisumu 40100, Kenya
Top