Volume 30, Number 8—August 2024
Research
Emergence of Bluetongue Virus Serotype 3, the Netherlands, September 2023
Abstract
Since 1998, notifiable bluetongue virus (BTV) serotypes 1–4, 6, 8, 9, 11, and 16 have been reported in Europe. In August 2006, a bluetongue (BT) outbreak caused by BTV serotype 8 began in northwestern Europe. The Netherlands was declared BT-free in February 2012, and annual monitoring continued. On September 3, 2023, typical BT clinical manifestations in sheep were notified to the Netherlands Food and Product Safety Consumer Authority. On September 6, we confirmed BTV infection through laboratory diagnosis; notifications of clinical signs in cattle were also reported. We determined the virus was serotype 3 by whole-genome sequencing. Retrospective analysis did not reveal BTV circulation earlier than September. The virus source and introduction route into the Netherlands remains unknown. Continuous monitoring and molecular diagnostic testing of livestock will be needed to determine virus spread, and new prevention strategies will be required to prevent BTV circulation within the Netherlands and Europe.
Bluetongue virus (BTV) is an arthropodborne virus that can cause clinical disease and death in ruminants. All ruminants are susceptible to infection by BTV; infections in New World camelids have also been described (1–3). Other species, including humans, are not susceptible to infection, indicating that bluetongue (BT) is not a zoonosis.
BTV is transmitted by Culicoides spp. biting midges and has been historically present only between latitudes 35°S and 50°N (4,5). The BTV serogroup consists of >30 serotypes, of which serotypes 1–24 are notifiable to the World Organisation for Animal Health (WOAH). Since 1998, notifiable BTV serotypes 1–4, 6, 8, 9, 11, and 16 and nonnotifiable BTV serotypes 25 and 27 have been found in Europe and the Mediterranean Basin (6). In 2006, bluetongue virus serotype 8 (BTV-8) emerged in northwestern Europe, and the Netherlands was the first country where the virus infection was detected (7,8). After a major BT outbreak caused by BTV-8 in Europe during 2006–2007, an emergency BTV-8 vaccine became available in 2008 (9,10). Many owners of cattle herds and small ruminant flocks participated in the voluntary vaccination program conducted by the government of the Netherlands (11), resulting in a dramatic decline in the number of BTV clinical notifications to the Netherlands Food and Consumer Product Safety Authority (NVWA) in 2008. At the end of 2008, BTV antibodies were found in >80% of the susceptible host populations tested because of either natural infection or vaccination. No new infections were observed after 2009, and, after 3 years of screening for possible BTV circulation, the Netherlands regained its official BT-free status in February 2012. This BT disease–free status has been monitored annually according to European Union (EU) regulation 1108/2008/EC and has been confirmed without interruption up to December 2022. However, because of the risk of introducing BTV-8 from neighboring countries, vaccination has been authorized and, therefore, some farmers still vaccinate their animals for BTV-8.
On September 3, 2023, clinical signs in sheep indicative of BT were notified to authorities in the Netherlands simultaneously by 2 veterinary practices located within the middle of the country. We describe the questions raised and actions taken during the first 3 weeks after the initial notified clinical case was confirmed as a BTV infection by laboratory diagnostic tests.
Sheep and Cattle Populations in the Netherlands and Clinical Examination
In 2022, the Netherlands had ≈1,080,631 sheep and ≈1,596,894 dairy cattle >2 years of age, distributed among ≈31,000 sheep farms and 14,000 cattle herds (12,13). Farms that notified authorities of possible BT were visited by a veterinary team, specializing in small ruminants, who reviewed reported clinical symptoms and collected samples for BT diagnosis. In addition, several farms already confirmed as BTV-positive were visited by Royal GD (formerly the Gezondheidsdienst voor Dieren) personnel, who clinically examined the sheep and cattle on those farms and described clinical signs.
Retrospective Study
We investigated whether the initial BT outbreak started in the area of the 4 BTV serotype 3 (BTV-3)–confirmed sheep farms in central Netherlands or whether the outbreak began earlier than September. We screened bulk tank cow milk samples submitted during August 2023 for routine testing from all over the Netherlands to determine if BTV antibodies were present. Royal GD coordinates a national monitoring program for which ≈12,000 (90%) dairy cattle farmers in the Netherlands submit monthly bulk tank milk samples. A total of 1,000 submitted milk samples were eligible for inclusion in the BTV screening.
We used identification and registration data from the Rijksdienst Voor Ondernemend Nederland (https://www.rvo.nl) to enable selection of dairy herd farms that did not purchase any cattle during the vector-active season (beginning in April 2023 until the start of the outbreak in September 2023) and only housed animals bred in the Netherlands. After applying the selection criteria, we were able to use bulk milk samples from 7,900 dairy herds for the screening. The Netherlands is divided into 20 compartments as proposed in the EU Commission Decision 2005/393/EC. We randomly selected 1,000 bulk milk samples to include all 20 compartments (Appendix Figure 1), resulting in ≈50 sampled herds per compartment and enabling a 14% prevalence estimate with 95% confidence. On September 11, we presented the first preliminary results to the government of the Netherlands. On September 13, additional data on vaccination purchases registered in the MediRund database (https://www.medirund.nl) during 2019 through September 2023 became available, and we combined those data with the results from the bulk milk screening.
BTV Genome Detection by PCR
We performed BTV-specific real-time reverse transcription PCR (RT-PCR) as previously described (14). In brief, we extracted virus RNA from 200 µL of EDTA blood by using the Magnapure 96 robotic machine (Roche, https://www.roche.com) and MagnaPure 96 DNA and Viral NA Small Volume Kit (Roche). For RT-PCR, we combined 5 μL eluted RNA and 15 μL LightCycler RNA Master HybProbe kit (Roche) reagent containing enzymes and BTV-specific primers and probe and loaded each reaction mixture per well into a 96-well plate. We amplified the resulting cDNA by using a LightCycler 480 II instrument (Roche) and LightCycler integrated software version 1.5.1, without the external predenaturation step (14).
Serologic Analysis using Competitive ELISA
We performed a competitive ELISA by using an ID Screen Bluetongue Competition ELISA kit (Innovative Diagnostics, https://www.innovative-diagnostics.com) according to the manufacturer’s protocol. This ELISA has a sensitivity and specificity of 100% for BTV-specific antibodies but cannot detect antibodies generated against the genetically related epizootic hemorrhagic disease virus. We measured optical density at 450 nm by using a Multiskan FC instrument (Thermo Fisher Scientific, https://www.thermofisher.com) and MikroWin software version 5.09 (Labsis, https://labsis.de) and calculated the percentage inhibition by using the positive and negative controls supplied with the ELISA kit.
Whole-Genome Sequencing
We extracted RNA from EDTA blood and amplified BTV genome segments by using a sequence-independent single-primer amplification approach. We performed first-strand cDNA synthesis by combining 5 µL RNA and Superscript III (Thermo Fisher Scientific) according to the manufacturer’s protocol and 2 μmol/L of the oligonucleotide 5′-GTTTCCCAGTCACGATA(N9)-3′. We incubated the mixture for 3 minutes at 95°C to denature double-stranded virus RNA, then cooled on ice. We added the remaining reagents and incubated the reaction at 25°C for 5 minutes, 42°C for 50 minutes, and 70°C for 15 minutes, and then stored at 4°C. We performed second-strand synthesis by using Sequenase (Thermo Fisher Scientific) according to the manufacturer’s protocol. We amplified the products by using Q5 high-fidelity DNA polymerase (New England Biolabs, https://www.neb.com) according to the manufacturer’s guidelines, 2 µmol/L of oligonucleotide 5′-GTTTCCCAGTCACGATA-3′, and the following cycle conditions: 94°C for 4 minutes; 68°C for 5 minutes; 35 cycles of 94°C for 30 seconds, 50°C for 1 minute, and 68°C for 3 minutes; then 68°C for 5 minutes and cooling at 10°C. To enhance the number of virus sequence reads, we performed a size selection of >200 bp by using SPRIselect beads (Beckman Coulter Life Sciences, https://www.beckman.com) at a ratio of 100 µL sample to 80 μL beads. We barcoded ≈150 ng cDNA from each sample by using the Native Barcoding Kit 96 V14 (Oxford Nanopore Technologies, https://www.nanoporetech.com) according to the manufacturer’s protocol and sequenced the cDNA by using a PromethION Flow Cell, R10 M version (Oxford Nanopore Technologies). To align the reads, we applied Minimap 2 version 2.26 (15) against a custom BTV reference database to construct a draft genome by using reference-based mapping. We deposited the sequences into GenBank on September 26, 2023 (accession nos. OR603992–4001).
We conducted phylogenetic analysis separately for each genome segment sequence by using BLAST (https://blast.ncbi.nlm.nih.gov) and the alignment results of the top 15 sequences used for the analysis. In addition, we added genome segment (Seg)-2 reference strains and a selected number of closely related BTV-3 strains to the phylogenetic analysis (16). We aligned the sequences by using MAFFT version 7.475 (17) and reconstructed the phylogeny by using maximum-likelihood analysis in IQ-TREE version 2.0.3 (18) and 1,000 ultrafast bootstrap replicates (19). We visualized the tree by using the ggtree R package (20).
Antibody Detection in Bulk Tank Milk by Indirect ELISA
For the retrospective analysis of BTV antibodies in bulk milk, we used the ID Screen Bluetongue Milk Indirect ELISA (Innovative Diagnostics) according to the manufacturer’s protocol; this ELISA uses the recombinant VP7 protein as the antigen and was validated in the Netherlands in 2007 (21). To study herd prevalence of BTV-3 infections in 2023, we used the cutoff values described in the ELISA manual: sample/positive control (S/P) values of <30% were considered negative, S/P values of >30% to <40% were considered potential positives, and S/P values >40% were considered positive. Using those cutoff values, the test had a sensitivity of ≈95% and specificity of 100%.
Geographic Distribution of BTV Clinical Cases
We graphically displayed sheep and cattle densities in thematic maps of the Netherlands according to 2-digit postal codes. We used BTV-confirmed clinical notifications of sheep and cattle cases until September 29, 2023. We generated all maps by using Stata version 17 (StataCorp LLC., https://www.stata.com).
Timeline of Outbreak
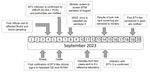
On September 3 and 4, 2023, NVWA was notified of clinical signs that were indicative of BTV infections at 5 sheep farms in the central region of the Netherlands near the Loosdrechtse Plassen. Flocks were visited by a team of veterinary specialists, and serum and EDTA blood samples were collected from the sheep and sent to the Netherlands National Veterinary Reference Laboratory for BTV at Wageningen Bioveterinary Research (WBVR).
On September 6, we confirmed BTV infections by real-time RT-PCR and competitive ELISAs (Figure 1). Of the 7 blood samples taken from 5 sheep farms, 6 samples from 4 different farms had BTV-positive RT-PCR results; cycle threshold values were 23–31. BTV antibodies were found in blood samples from 5 of the 6 PCR-positive sheep; blocking was >90% in competitive ELISAs. We immediately reported the findings to the Ministry of Agriculture, Nature and Food Quality of the Netherlands, and requested new samples for confirmation and shipment to the EU Reference Laboratory for BTV, Center for Animal Health Research, National Center for Agricultural and Food Research and Technology, in Madrid, Spain. In addition, using nanopore technology, WBVR conducted whole-genome sequencing on the RT-PCR–positive samples. The first suspicion of BTV in cattle was also notified to the NVWA on September 6.
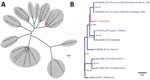
On September 8, three blood samples from sheep sampled at 3 unrelated farms showed sufficient sequence coverage per nucleotide (range 30–2,570 reads) (Table 1) to reliably determine contig sequences for all 10 genome segments for those 3 samples. Contig sequences derived from individual samples were 100% identical. Contigs represented full-length sequences of Seg-1–Seg-9, including the 5′ and 3′ termini. Contigs of Seg-10 were incomplete and were completed by using Sanger sequencing, except for the ultimate 22 nt at the 3′ end, corresponding to the amplification primer. Phylogenetic analysis of Seg-2, which encodes the serotype-dominant VP2 protein, along with prototypic isolates of WOAH-notifiable BTV serotypes 1–24 identified the causative BT agent as BTV-3, which was designated as variant BTV-3/NET2023. Phylogenetic clustering of sequences was also observed with serotypes 13 and 16, confirming previous genetic analysis (Figure 2, panel A) (22). On the basis of the phylogenetic tree, WBVR announced that genotyping revealed the BT outbreak was caused by BTV-3 because of the high homology with known serotype 3 isolates. Detailed phylogenetic analysis showed a close relationship with Seg-2 from BTV-3 isolates from Italy and Tunisia (Figure 2, panel B). Phylogenetic analyses of other genome segments of BTV-3/NET2023 did not indicate a particular ancestor but had close identity (>97%) to genome segments of various other sequenced BTV isolates deposited in GenBank (Table 2).
On September 11, WBVR confirmed BTV in newly collected serum and EDTA blood samples from all 4 initially infected sheep farms and sent the samples to the EU Reference Laboratory for confirmation and serotyping by serotype-specific real-time RT-PCR. On September 14, the EU Reference Laboratory confirmed the results by using the WOAH-recommended RT-PCR test targeting Seg-10. RT-PCR specific for serotypes 3, 4, and 8 clearly confirmed serotype 3, and the results were immediately forwarded to the Ministry of Agriculture, Nature and Food Quality of the Netherlands and NVWA.
On September 19, the first suspicion of BTV in a goat was notified to the NVWA; specimens collected from 1 goat were positive for BTV by real-time RT-PCR. In addition, BTV-3/NET2023 isolation from sheep EDTA blood from the initial 4 farms was successful by using Culicoides-derived KC cells (23).
Clinical Manifestations in Sheep, Cattle, and Goats
BTV-3–infected sheep showed signs of fever, lethargy, hypersalivation, ulcerations and erosions of the oral and nasal mucous membranes, facial edema, lesions of the coronary band, lameness, and death (Figure 3). Several days after the initial outbreak confirmation in sheep, clinical signs were also reported in cattle. Clinical signs observed in cattle were fever, apathy, conjunctivitis, nasal discharge, erosions and crust formation on lips and nostrils, ulcerations and erosions of oral mucosa, edema of the nose, coronitis, and superficial necrosis of teats. The goat reported on September 19 showed signs of edema of the lips and fever (Appendix Figure 2).
After the outbreak began, the number of notifications increased rapidly for both sheep flocks and cattle herds (Figure 4). The initial cases included 4 sheep farms. One week later (calendar week 36), a total of 25 sheep flock and 12 cattle herd notifications were confirmed as BTV-3–positive by RT-PCR. In the second week (calendar week 37), the total number of positive suspicions increased to 18 sheep flocks and 55 cattle herds. In the third week (calendar week 38), the total number of PCR-positive BTV-3–diagnosed flocks or herds increased to 324 sheep flocks, 61 cattle herds, and 1 goat herd.
Retrospective Study
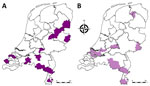
To investigate whether the BT outbreak began at the 4 initially-infected sheep farms, we screened bulk tank milk samples submitted in August from cattle farms for routine BTV antibody testing. Of the 991 bulk tank milk samples, 955 tested negative, 8 tested potentially positive, and 28 tested positive for BTV antibodies; antibody prevalence was 2.8% (95% CI 1.9%–4.1%) (Table 3). However, 24 of 36 bulk tank milk samples showing potentially positive or positive results (67%) were from farms that had a proven history of animal vaccination against BTV-8. BTV antibodies that could not be linked to a recorded history of BTV vaccination were found in 12 of 991 bulk tank milk samples. However, the 12 antibody-positive herds were not clustered, and 7 of those 12 herds were located near the borders with Belgium and Germany, suggesting a high likelihood that those farms in the Netherlands might have vaccinated their animals because of the presence of BTV-8 in Belgium and Germany (Figure 5). Altogether, no area in the Netherlands had a high seroprevalence for BTV antibodies in August 2023 according to cow milk sampling, and no BTV-specific antibodies were found in the region where the initial BTV notifications were made. Therefore, the 4 BT index cases occurred in the 4 initially affected sheep farms.
We describe the actions taken after a novel BTV-3 strain emerged in sheep and cattle in the Netherlands. Initially, sheep from 4 farms showed clinical signs of fever, lethargy, hypersalivation, ulcerations, erosions of the oral and nasal mucous membranes, or sudden death. The sheep were positive for BTV by real-time RT-PCR, and all but 1 showed seroconversion by using a competition ELISA. Whole-genome sequencing using nanopore technology showed the full virus genome sequence could be characterized quickly, and the generated nucleotide sequence of Seg-2 aligned with other BTV-3 sequences. We investigated BTV epidemiology during the month before the first cases were reported in sheep by retrospectively testing bulk tank cow milk; however, either no high seroprevalence was observed or seropositive samples were found within the region where the initial cases were detected. A very low number of antibody-positive bulk milk samples (n = 12) were found that could not be linked to previous vaccination. It is possible that the cows from those herds were still vaccinated, but that possibility could not be substantiated on the basis of available data. In addition, those findings might have been false positives despite the high specificity of the ELISA (21). We concluded that a massive spread of BTV did not occur before the first detection of BTV-3 in the sheep farms, which agrees with the findings of a retrospective analysis of 1,003 sheep serum samples from 89 flocks that indicated a 3.4% BTV herd prevalence in August (I.M.G.A. Santman-Berends et al., unpub. data). As of March 12, 2024, animals from a total of 4,371 farms or holdings have been confirmed as BTV-3 positive by real-time PCR.
Early detection of diseases by clinical diagnosis remains challenging, especially for unpredicted nonendemic diseases. BT displays a wide and nonspecific spectrum of clinical manifestations in ruminants, such as fever, hypersalivation, lameness, edema, and sudden death. BT disease severity in sheep and cattle overlaps with several other endemic infections, such as orf, dermatophilosis, haemonchosis, pasteurellosis, strawberry footrot, and photosensitization, which are relevant, differentially diagnosed endemic conditions in sheep. Malignant catarrhal fever and photosensitization can cause signs similar to BT in cattle (24,25). Awareness of BTV-like symptoms by veterinarians is also of great importance for other notifiable diseases, such as foot-and-mouth disease, peste des petits ruminants, sheep and goat pox, and epizootic hemorrhagic disease, and should be notified to the official authorities when suspected.
During a BT outbreak, communication creates increased awareness among veterinarians and farmers, which might lead to an increase in false BT notifications because of nonspecific clinical signs. Therefore, education and training of veterinarians and livestock farmers about the clinical manifestations of BT and other diseases remains critical, especially for notifiable diseases that have not occurred for an extended time, because many veterinarians might not have seen the clinical symptoms in their practice. Only a laboratory diagnosis can and should rapidly differentiate between notifiable diseases to support a clinical diagnosis. Nevertheless, in the outbreak described in this study, the emerging BT disease was detected successfully at an early stage.
The rapid spread of BTV after the initial emergence shows that indigenous Culicoides spp. midges in the Netherlands are competent vectors for transmitting BTV-3/NET2023. The BTV vector, the C. imicola midge, found predominantly in Africa and Asia, is not found in northwestern Europe, and BTV-6 introduction in the Netherlands in 2008 showed that the outbreak dies out when indigenous Culicoides spp. midges are unable to effectively transmit the virus (26). BTV-3/NET2023 is the second BTV variant, after BTV-8/NET2006, that has been successfully transmitted by indigenous biting midge species in northwestern Europe (27). BTV-8/NET2006 is transmitted by indigenous biting midge species of the C. obsoletus complex, including C. obsoletus, C. scoticus, C. dewulfi, and C. chiopterus (28–30). Entomologic research is needed to identify the biting midge species involved in BTV-3/NET2023 transmission.
The geographic origin and route of introduction of BTV-3/NET2023 into the Netherlands are unknown. Phylogenetic analysis of Seg-2 shows clustering with other BTV-3 Seg-2 sequences, including geographically close BTV-3 variants. However, BTV-3 has only been described in Europe in Sicily and Sardinia, Italy, and the few sequences available from those virus isolates show a relatively high variation compared with BTV-3/NET2023 sequences from the Netherlands. In addition, other genome segment sequences did not show high homology with sequences from different BTV-3 variants. Therefore, tracing the origin of BTV-3/NET2023 has been difficult. The segmented genome of BTV enables reassortment, also known as antigenic shift, between variants, which further hampers unravelling the geographic source of BTV-3/NET2023. The virus was likely introduced into the Netherlands from a distant source because the neighboring countries Belgium and Germany have had BT-free status since June 2023. Although the source and route of BTV-3/NET2023 is unclear, yearly monitoring and the findings from this retrospective study indicate that virus circulation began in September 2023 within the Netherlands.
In conclusion, after a decade of having BT-free status, the Netherlands saw BTV-3 emerge in 2023, causing clinical signs and death in sheep and cattle. The causative virus strain is designated BTV-3/NET2023. The source, geographic origin, and introduction route of BTV-3/NET2023 are unknown, but virus circulation has rapidly expanded. BTV-3/NET2023 is transmitted by indigenous biting midges, but the vector-competent midge species has not yet been identified. Continuous monitoring and molecular diagnostic testing of sheep, cattle, and goats will be needed to determine virus spread, and new vaccination and other prevention strategies will be required to prevent BTV circulation within the Netherlands and Europe.
Dr. Holwerda is head of the national reference laboratory for viral veterinary vectorborne diseases at Wageningen Bioveterinary Research. His research interests focus on the diagnosis and pathogenesis of veterinary virus infections.
Acknowledgments
We thank the veterinary practitioners from Dierenartsenpraktijk Gorter and Dierenkliniek Amstel in Vecht en Venen, all the laboratory personnel at WBVR and Royal GD, and all veterinarians who conducted the herd visits for their laborious work and for providing photographs.
This study was supported by the Netherlands Ministry of Agriculture, Nature and Food Quality (project no. 1600002757 VZVD).
References
- Henrich M, Reinacher M, Hamann HP. Lethal bluetongue virus infection in an alpaca. Vet Rec. 2007;161:764. DOIPubMedGoogle Scholar
- Meyer G, Lacroux C, Léger S, Top S, Goyeau K, Deplanche M, et al. Lethal bluetongue virus serotype 1 infection in llamas. Emerg Infect Dis. 2009;15:608–10. DOIPubMedGoogle Scholar
- Ortega J, Crossley B, Dechant JE, Drew CP, Maclachlan NJ. Fatal bluetongue virus infection in an alpaca (Vicugna pacos) in California. J Vet Diagn Invest. 2010;22:134–6. DOIPubMedGoogle Scholar
- Zhang N, MacLachlan NJ, Bonneau KR, Zhu J, Li Z, Zhang K, et al. Identification of seven serotypes of bluetongue virus from the People’s Republic of China. Vet Rec. 1999;145:427–9. DOIPubMedGoogle Scholar
- Mellor PS, Carpenter S, Harrup L, Baylis M, Mertens PPC. Bluetongue in Europe and the Mediterranean Basin: history of occurrence prior to 2006. Prev Vet Med. 2008;87:4–20. DOIPubMedGoogle Scholar
- Lundervold M, Milner-Gulland EJ, O’Callaghan CJ, Hamblin C. First evidence of bluetongue virus in Kazakhstan. Vet Microbiol. 2003;92:281–7. DOIPubMedGoogle Scholar
- van Wuijckhuise L, Dercksen D, Muskens J, de Bruijn J, Scheepers M, Vrouenraets R. [Bluetongue in The Netherlands; description of the first clinical cases and differential diagnosis. Common symptoms just a little different and in too many herds] [in Dutch]. Tijdschr Diergeneeskd. 2006;131:649–54.PubMedGoogle Scholar
- Elbers ARW, Backx A, Meroc E, Gerbier G, Staubach C, Hendrickx G, et al. Field observations during the bluetongue serotype 8 epidemic in 2006. I. Detection of first outbreaks and clinical signs in sheep and cattle in Belgium, France and the Netherlands. Prev Vet Med. 2008;87:21–30. DOIPubMedGoogle Scholar
- Wäckerlin R, Eschbaumer M, König P, Hoffmann B, Beer M. Evaluation of humoral response and protective efficacy of three inactivated vaccines against bluetongue virus serotype 8 one year after vaccination of sheep and cattle. Vaccine. 2010;28:4348–55. DOIPubMedGoogle Scholar
- Oura CAL, Edwards L, Batten CA. Evaluation of the humoral immune response in adult dairy cattle three years after vaccination with a bluetongue serotype 8 inactivated vaccine. Vaccine. 2012;30:112–5. DOIPubMedGoogle Scholar
- Elbers ARW, de Koeijer AA, Scolamacchia F, van Rijn PA. Questionnaire survey about the motives of commercial livestock farmers and hobby holders to vaccinate their animals against Bluetongue virus serotype 8 in 2008-2009 in the Netherlands. Vaccine. 2010;28:2473–81. DOIPubMedGoogle Scholar
- Dijkstra E, Vellema P, Peterson K, Bogt-Kappert CT, Dijkman R, Harkema L, et al. Monitoring and surveillance of small ruminant health in the Netherlands. Pathogens. 2022;11:635. DOIPubMedGoogle Scholar
- Santman-Berends IMGA, Brouwer-Middelesch H, Van Wuijckhuise L, de Bont-Smolenaars AJG, Van Schaik G. Surveillance of cattle health in the Netherlands: Monitoring trends and developments using routinely collected cattle census data. Prev Vet Med. 2016;134:103–12. DOIPubMedGoogle Scholar
- van Rijn PA, Heutink RG, Boonstra J, Kramps HA, van Gennip RGP. Sustained high-throughput polymerase chain reaction diagnostics during the European epidemic of Bluetongue virus serotype 8. J Vet Diagn Invest. 2012;24:469–78. DOIPubMedGoogle Scholar
- Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34:3094–100. DOIPubMedGoogle Scholar
- Maan S, Maan NS, Samuel AR, Rao S, Attoui H, Mertens PPC. Analysis and phylogenetic comparisons of full-length VP2 genes of the 24 bluetongue virus serotypes. J Gen Virol. 2007;88:621–30. DOIPubMedGoogle Scholar
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–80. DOIPubMedGoogle Scholar
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–74. DOIPubMedGoogle Scholar
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 2018;35:518–22. DOIPubMedGoogle Scholar
- Yu G, Smith DK, Zhu H, Guan Y, Lam TTY. GGTREE: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol. 2017;8:28–36. DOIGoogle Scholar
- Mars MH, van Maanen C, Vellema P, Kramps JA, van Rijn PA. Evaluation of an indirect ELISA for detection of antibodies in bulk milk against bluetongue virus infections in the Netherlands. Vet Microbiol. 2010;146:209–14. DOIPubMedGoogle Scholar
- van Rijn PA. Prospects of next-generation vaccines for bluetongue. Front Vet Sci. 2019;6:407. DOIPubMedGoogle Scholar
- Wechsler SJ, McHolland LE, Wilson WC. A RNA virus in cells from Culicoides variipennis. J Invertebr Pathol. 1991;57:200–5. DOIPubMedGoogle Scholar
- Williamson S, Woodger N, Darpel K. Differential diagnosis of bluetongue in cattle and sheep. In Pract. 2008;30:242–51. DOIGoogle Scholar
- Dercksen D, Lewis C. Bluetongue virus serotype 8 in sheep and cattle: a clinical update. In Pract. 2007;29:314–8. DOIGoogle Scholar
- van Rijn PA, Geurts Y, van der Spek AN, Veldman D, van Gennip RGP. Bluetongue virus serotype 6 in Europe in 2008-Emergence and disappearance of an unexpected non-virulent BTV. Vet Microbiol. 2012;158:23–32. DOIPubMedGoogle Scholar
- Mellor PS, Boorman J, Baylis M. Culicoides biting midges: their role as arbovirus vectors. Annu Rev Entomol. 2000;45:307–40. DOIPubMedGoogle Scholar
- Mehlhorn H, Walldorf V, Klimpel S, Jahn B, Jaeger F, Eschweiler J, et al. First occurrence of Culicoides obsoletus-transmitted Bluetongue virus epidemic in Central Europe. Parasitol Res. 2007;101:219–28. DOIPubMedGoogle Scholar
- Meiswinkel R, van Rijn P, Leijs P, Goffredo M. Potential new Culicoides vector of bluetongue virus in northern Europe. Vet Rec. 2007;161:564–5. DOIPubMedGoogle Scholar
- Dijkstra E, van der Ven IJK, Meiswinkel R, Hölzel DR, Van Rijn PA, Meiswinkel R. Culicoides chiopterus as a potential vector of bluetongue virus in Europe. Vet Rec. 2008;162:422. DOIPubMedGoogle Scholar
Figures
Tables
Cite This ArticleOriginal Publication Date: June 28, 2024
1These authors contributed equally to this article.
Table of Contents – Volume 30, Number 8—August 2024
| EID Search Options |
|---|
|
|
|
|
|
|


Please use the form below to submit correspondence to the authors or contact them at the following address:
Melle Holwerda, Wageningen Bioveterinary Research (WBVR), Department of Virology, PO Box 65, 8200 AB Lelystad, the Netherlands
Top