Standardized Phylogenetic Classification of Human Respiratory Syncytial Virus below the Subgroup Level
Stephanie Goya
1
, Christopher Ruis
2, Richard A. Neher
2, Adam Meijer, Ammar Aziz, Angie S. Hinrichs, Anne von Gottberg, Cornelius Roemer, Daniel G. Amoako
3, Dolores Acuña, Jakob McBroome, James R. Otieno, Jinal N. Bhiman, Josie Everatt, Juan C. Muñoz-Escalante, Kaat Ramaekers
4, Kate Duggan, Lance D. Presser, Laura Urbanska, Marietjie Venter, Nicole Wolter, Teresa C.T. Peret, Vahid Salimi, Varsha Potdar, Vítor Borges, and Mariana Viegas
1
Author affiliations: University of Washington, Seattle, Washington, USA (S. Goya); University of Cambridge, Cambridge, UK (C. Ruis); University of Basel and SIB, Basel, Switzerland (R.A. Neher, C. Roemer, L. Urbanska); National Institute for Public Health and the Environment, Bilthoven, the Netherlands (A. Meijer, L.D. Presser); World Health Organization Collaborating Centre for Reference and Research on Influenza, Melbourne, Victoria, Australia (A. Aziz); University of California Santa Cruz, Santa Cruz, California, USA (A.S. Hinrichs, J. McBroome); National Institute for Communicable Diseases of the National Health Laboratory Service, Johannesburg, South Africa (A. von Gottberg, J.N. Bhiman, J. Everatt, N. Wolter); University of Witwatersrand, Johannesburg, South Africa (A. von Gottberg, J.N. Bhiman, N. Wolter); University of KwaZulu-Natal, Durban, South Africa (D.G. Amoako); Universidad Nacional de La Plata, Buenos Aires, Argentina (D. Acuña, M. Viegas); National Scientific and Technical Research Council, Buenos Aires, Argentina (D. Acuña, M. Viegas); Theiagen Genomics, Highlands Ranch, Colorado, USA (J.R. Otieno); Autonomous University of San Luis Potosí, San Luis Potosí, Mexico (J.C. Muñoz-Escalante); Rega Institute for Medical Research, Leuven, Belgium (K. Ramaekers); University of Edinburgh, Edinburgh, Scotland, UK (K. Duggan); University of Pretoria, Pretoria, South Africa (M. Venter); University of Texas Medical Branch, Galveston, Texas, USA (T.C.T. Peret); Tehran University of Medical Sciences, Tehran, Iran (V. Salimi); ICMR National Institute of Virology, Pune, India (V. Potdar); National Institute of Health Doutor Ricardo Jorge, Lisbon, Portugal (V. Borges)
Main Article
Figure 4
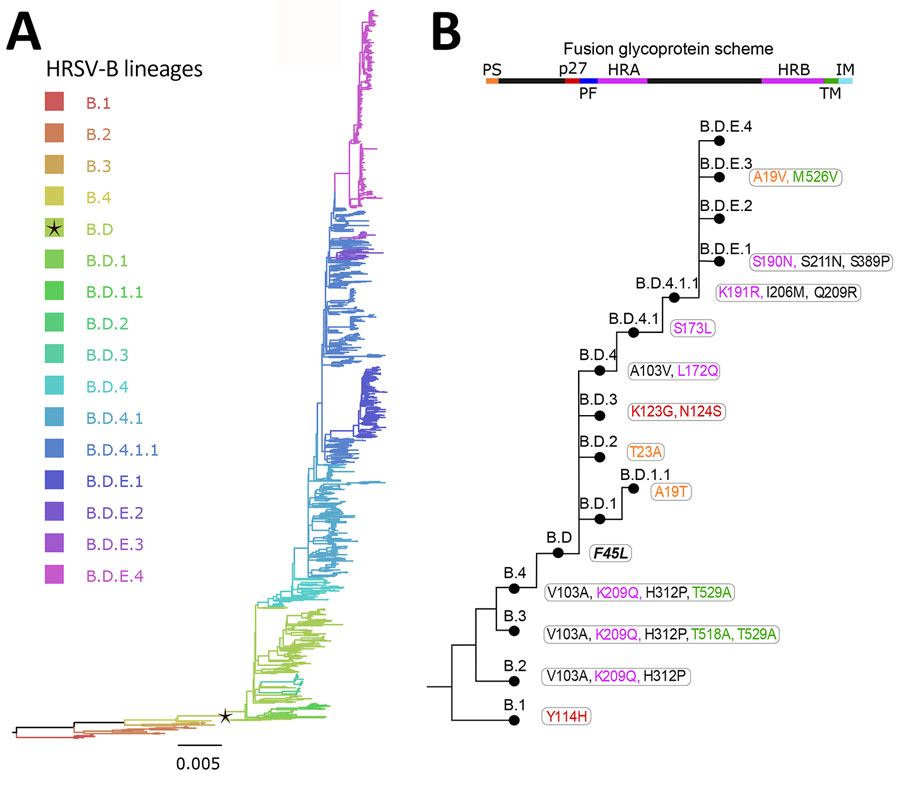
Figure 4. Human respiratory syncytial virus B lineages classification. A) HRSV-B maximum-likelihood phylogenetic tree (1,385 sequences), colored according to lineage classification. Black star indicates B.D lineage, defined by the 60-nt duplication in the G gene. Scale bar indicates substitutions per site. B) Simplified scheme of the lineage designation to highlight the presence of nested lineages. The amino acid changes in the F glycoprotein are listed next to lineage name and colored according to their location in the fusion protein.
Main Article
Page created: June 21, 2024
Page updated: July 20, 2024
Page reviewed: July 20, 2024
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
