Disclaimer: Early release articles are not considered as final versions. Any changes will be reflected in the online version in the month the article is officially released.
Volume 31, Number 2—February 2025
Research
Epidemiologic and Genomic Surveillance of Vibrio cholerae and Effectiveness of Single-Dose Oral Cholera Vaccine, Democratic Republic of the Congo
Figure 4
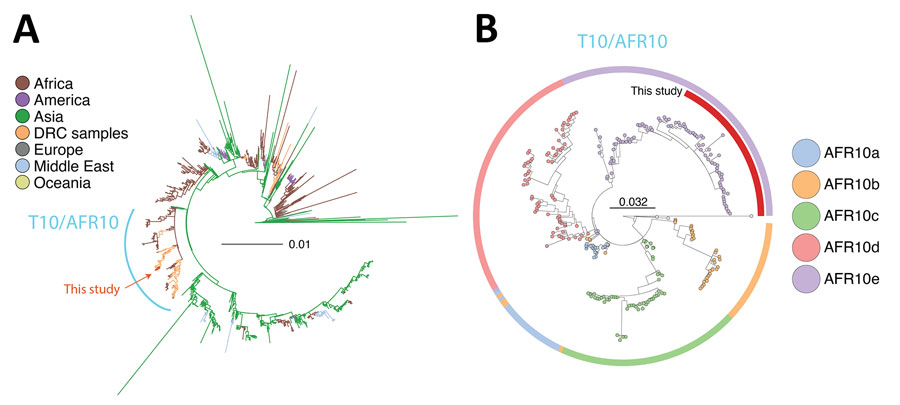
Figure 4. Phylogenetic analysis of Vibrio cholerae strains in study of V. cholerae transmission and effectiveness of single-dose oral cholera vaccine, DRC. Maximum-likelihood phylogenetic trees were prepared to compare V. cholerae seventh pandemic El Tor (7PET) isolates. A) Globally representative phylogeny of 1,428 7PET strains; 5 representative isolates from this study (red) were placed within the larger context of those 7PET strains. Tree was rooted on the A6 strain. Branch colors indicate geographic origin of the strain. B) Phylogeny of the T10/AFR10 lineage of V. cholerae. Colors indicate different V. cholerae lineages. Representative isolates from this study (n = 46; red) were placed within the context of 221 T10/AFR10 lineage strains. Tree was rooted on the reference strain N16961. Scale bars indicate nucleotide substitutions per site. DRC, Democratic Republic of the Congo.
1These first authors contributed equally to this article.