Disclaimer: Early release articles are not considered as final versions. Any changes will be reflected in the online version in the month the article is officially released.
Volume 31, Number 5—May 2025
Research Letter
Molecular Epidemiology of St. Louis Encephalitis Virus, São Paulo State, Brazil, 2016–2018
Figure
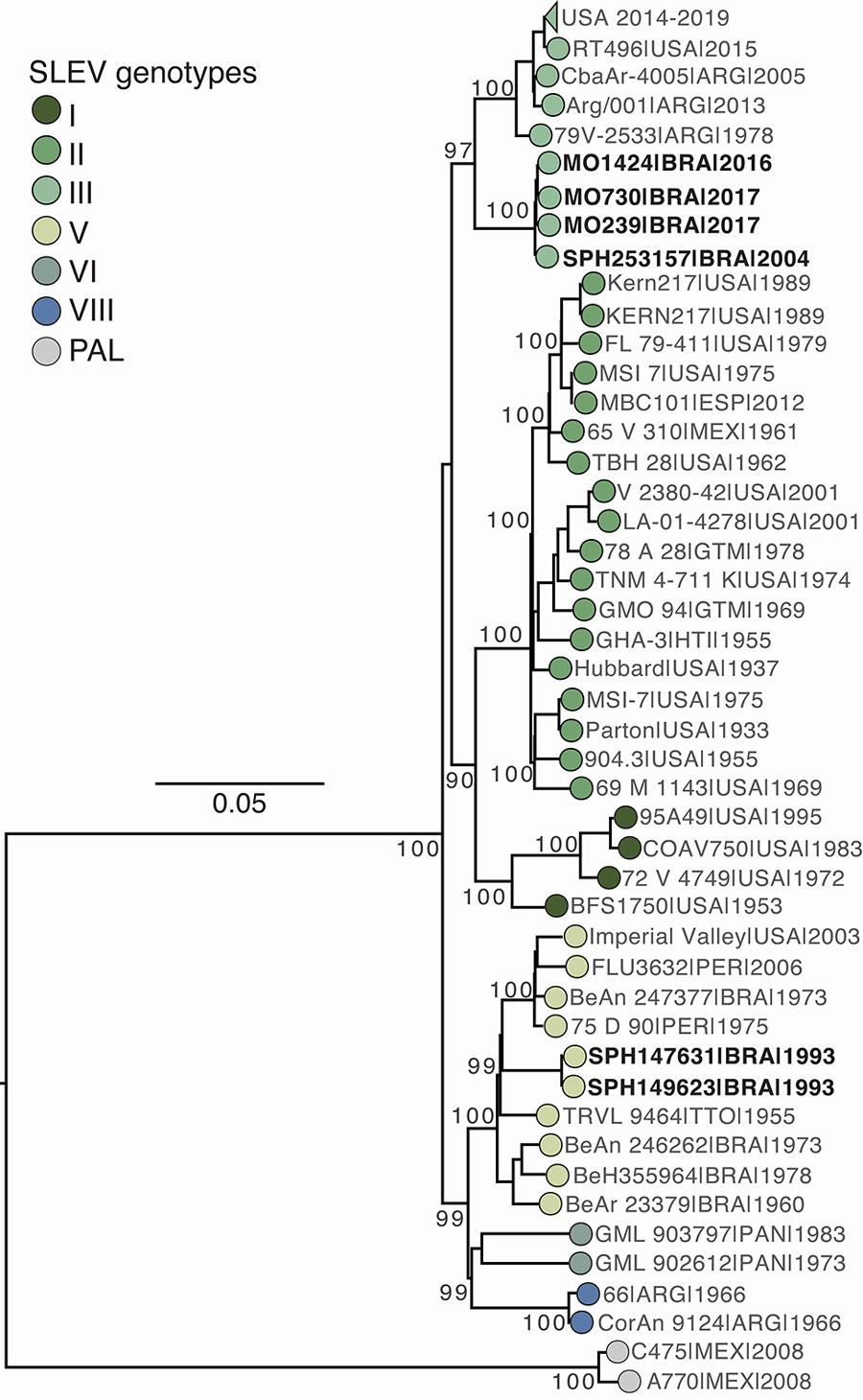
Figure. Maximum-likelihood phylogenetic tree in a study of molecular epidemiology of St. Louis encephalitis virus in São Paulo State, Brazil, 2016–2018. Tree shows 219 representative SLEV complete coding sequence genomes, including 3 new genomes generated in this study from Aedes albopictus (MO239), Ae. aegypti (MO1424), and Sabethes chloropterus (MO730) mosquitoes. The tree also includes 3 historical partially sequenced or unsequenced SLEV isolates from São Paulo state: SPAR149623 (Culex sp. mosquito, Santo Antônio de Aracanguá, May 12, 1993), SPAR147631 (Anopheles triannulatus mosquito, Pereira Barreto, March 11, 1993), and SPH253157 (human, São Pedro, January 1, 2004). Tree tips are colored by genotype. Phylogenies are midpoint-rooted for clarity; bootstrap values (1,000 replicates) are shown on major nodes. Scale bar represents nucleotide substitutions per site. GenBank accession numbers for the sequences and detailed information on the collapsed USA clade (genotype III, circulating during 2014–2019) is provided in the Appendix. SLEV, St. Louis encephalitis virus.
1These authors contributed equally to this article.