Volume 9, Number 2—February 2003
Dispatch
Invasive Type e Haemophilus influenzae Disease in Italy
Abstract
We describe the first reported cases of invasive type e Haemophilus influenzae disease in Italy. All five cases occurred in adults. The isolates were susceptible to ampicillin and eight other antimicrobial agents. Molecular analysis showed two distinct type e strains circulating in Italy, both containing a single copy of the capsulation locus.
In Italy, the vaccine against Haemophilus influenzae type b (Hib) was licensed in 1995, but vaccination is voluntary. Vaccination coverage by 24 months of age was estimated in 1998 at 19.8% for the 1996 birth cohort (1). Coverage increased in 2000 to 53.1% for the 1998 birth cohort (2). Since 1994, surveillance of H. influenzae meningitis has been conducted within the National Surveillance of Bacterial Meningitis (3). A laboratory-based active surveillance of invasive H. influenzae disease was implemented in a sample of Italian regions in 1997 (4). In 1997–1998, the incidence of invasive Hib disease estimated by this system was lower than that reported in northern and central Europe and in the United States before mass vaccination was introduced, yet was comparable with the incidence reported in other Mediterranean countries with similar vaccination coverage (4).
In 1998–2001, laboratory-based active surveillance was conducted in seven Italian regions, including a population of approximately 24 million persons (33% of the Italian population). Participating regions were located throughout the country and included rural, urban, and large metropolitan areas. Active H. influenzae case finding was conducted by contacting, monthly, microbiologists from the regional laboratories of hospitals with infectious disease or pediatric wards. A patient with invasive disease was defined as a patient with a compatible illness, accompanied by isolation of H. influenzae from a normally sterile site or detection of Hib antigen in cerebrospinal fluid. Hospital microbiologists were asked to send H. influenzae isolates to the national reference laboratory at Istituto Superiore di Sanità, where all strains were assayed by polymerase chain reaction (PCR) capsular genotyping. Serotyping by slide agglutination was performed at the regional level, when possible, and at the national reference laboratory.
From 1998 to 2001, a total of 219 cases of invasive H. influenzae disease were reported; 165 were diagnosed by isolation of H. influenzae from a normally sterile site, and 54 were diagnosed by detection of Hib antigen in cerebrospinal fluid. Of the 165 isolates, 97 (58.8%) were sent to the national reference laboratory; the percentage of isolates sent to the reference laboratory remained relatively stable over the years (from 62.3% in 1998 to 58.3% in 2001). Analysis of incidence data by serotype showed that the annual number of cases of invasive Hib disease decreased from 69 in 1998 to 17 in 2001, while the number of nontypable H. influenzae remained constant (mean 8 cases/year; range 7–11) (Table 1). No cases attributable to capsulated H. influenzae other than type b were detected in years 1998–1999; in the next 2 years, a total of seven cases were traced; five were due to type e strain.
We describe these five cases of invasive disease caused by H. influenzae type e (Hie). Genetic relationship among the five Hie isolates was assessed by pulsed-field gel electrophoresis (PFGE). Susceptibility to nine antimicrobial agents, including ampicillin, was also determined. Because amplification of capsulation (cap) locus is assumed to contribute to strain virulence, the copy number of cap e locus in each isolate was also identified.
Five Hie isolates were detected through surveillance from January 2000 to December 2001. Characteristics of patients were obtained by reviewing clinical records (Table 2). Briefly, two cases, both with meningitis, occurred in young adults, who recovered. The remaining three cases, two with bacteremic pneumonia and one with septicemia, were in elderly patients, who died. All but one (no. 2) of the patients were from neighboring regions in northeastern Italy. However, the towns they lived in were quite distant, and the patients were admitted to different hospitals in different periods. Serotyping of the isolates by slide agglutination was performed by using polyvalent and monovalent antisera to capsular serotypes a through f (Difco Laboratories, Detroit, MI). Capsular genotype was identified by PCR (5). Briefly, in a first round of PCR, primers to the ompP2 gene were used to confirm the H. influenzae species, while primers directed to the bex region confirmed capsulation. A second round of PCR with primers directed to the cap e–specific region (6) generated an expected product of 1,350 bp. Further characterization of the isolates was performed by phenotypic and genotypic methods. Biotypes were assigned by determining indole production, urease level, and ornithine decarboxylase activities. MICs of ampicillin were determined by E-test (AB Biodisk, Solna, Sweden). Hib strain ATCC 10211 was used as control. Susceptibilities to trimethoprim/sulfamethoxazole, ceftazidime, chloramphenicol, azithromycin, aztreonam, ciprofloxacin, imipenem, and tetracycline were tested by disk diffusion assay. Both the E-test and the disk diffusion assay were performed by using Haemophilus test medium, as recommended by the National Committee for Clinical Laboratory Standards (NCCLS) (7). Interpretative breakpoints and zone diameters were also based on NCCLS criteria. Production of β-lactamase was detected by the cefinase disk test (BBL, Becton-Dickinson, Sparks, MD). Restriction fragment length polymorphism analysis by PFGE was conducted as described (5), except for digestion of genomic DNA, performed by using SmaI (20 U) or ApaI (20 U) restriction enzymes (New England BioLabs, Wilbury Way Hitchin, U.K.). The copy number of cap e locus was determined by Southern blot analysis, using as probe the 1,350-bp amplicon obtained by PCR of a prototype type e strain. Since the KpnI and SmaI sites flank the cap locus of encapsulated H. influenzae strains, the copy number of the locus can be estimated by the size of the restriction fragment obtained after digestion of the chromosome with these enzymes (8). Restriction fragments were separated by PFGE as described (5), transferred to nylon membranes, and hybridized with the probe. Labeling of the probe and hybridization reactions were obtained by using the ECL kit (Amersham Pharmacia Biotech, Little Chalfont, U.K.). Hybridizing bands were visualized by autoradiographs. Although no data are available on the size of cap e locus, on the basis of results obtained on Hib strains, the DNA fragment for a single-copy strain was expected to be approximately 27–28 kb. In fact, the KpnI/SmaI fragment includes the cap locus, whose size is 18 kb in Hib strains, plus additional segments (about 10 kb) upstream and downstream of the cap region (8). Strains with two or more copies of the cap b locus featured fragments of increased size (45 kb, 63 kb, 81 kb, 99 kb) (8).
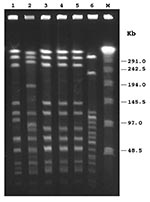
By PCR, all isolates exhibited the type e capsular genotype. By slide agglutination method, performed at the national reference laboratory, four isolates were designed as type e, and one was misidentified as nontypable. At the regional level, only one of the five PCR-positive strains had been recognized as type e, two had been identified as nontypable, and two had not been typed. By biotyping, four isolates were classified as biotype IV, and one as biotype I (patient no. 5) (Table 2). MICs of ampicillin ranged from 0.125 µg/mL to 0.25 µg/mL, indicating that all isolates were susceptible to this antibiotic; none produced β-lactamase. As assessed by disk diffusion assay, all isolates were also susceptible to trimethoprim/sulfamethoxazole, ceftazidime, chloramphenicol, azithromycin, aztreonam, ciprofloxacin, imipenem, and tetracycline. PFGE with SmaI restriction enzyme digestion generated not well-resolved profiles, as several very close fragments of 194–145 kb were obtained (data not shown). Following ApaI digestion, profiles were easier to compare (Figure): three isolates (patients no. 3, 4, and 5) shared an indistinguishable pattern (pattern 1); one isolate (patient no. 1) showed a profile closely related to pattern 1 but with two band differences (pattern 1a). The isolate from patient no. 2 appeared clearly different from the others (pattern 2), according to criteria reported by Tenover et al. (10) (Table 2). All Hie isolates were unrelated to invasive Hib strains circulating in Italy (9) (Figure). According to epidemiologic data, patients with strains that showed an indistinguishable PFGE pattern did not appear to share any common risk factor. Southern hybridization with the probe for the cap e gene after PFGE identified a single fragment of 20.5 kb in each isolate tested, suggesting the presence of one copy of the cap locus (Table 2).
Although studies on nasopharyngeal carriage of H. influenzae have shown the presence of serotype e as colonizer (11–13), few studies have described serious infections attributable to this capsular type (14–17). Our results suggest that Hie may cause either fatal infections in elderly patients with underlying disease or meningitis in adults whether or not underlying conditions are present. In Italy, these cases of Hie infection are the first reported. Despite limited data on the frequency of non–type b invasive disease before 1997, our results are based on 4 years of laboratory-based active surveillance on a large population sample. Since no type e strains were detected in the first 2 years of this surveillance, this clustering is likely due to the emergence of invasive Hie disease. Alternative explanations might include improved laboratory confirmation methods (however, the proportion of meningitis cases without an identified etiologic agent did not decrease from 1998 to 2001) or improved reporting in the last 2 years of surveillance (however, no changes to the system were made, and quality indicators, such as proportion of isolates sent to the reference laboratory, remained stable throughout the 4 years).
An unequivocal method of assigning capsular type is required to monitor infection attributable to uncommon serotypes, especially since, as in some Hie isolates, expression of capsular polysaccharide is not sufficient to be detected by slide agglutination (18). In our study, PCR capsular typing allowed the identification of an additional Hie strain that had been misidentified by slide agglutination.
The prevalence of ampicillin resistance in both β-lactamase–positive and –negative H. influenzae isolates has increased worldwide. Neither β-lactamase production nor intrinsic resistance was detected among our Hie isolates, confirming the low incidence of ampicillin-resistant H. influenzae strains from invasive disease in Italy (5,9).
PFGE has been successfully used to type H. influenzae isolates. Our results suggest that two distinct Hie strains circulated in Italy in 2000–2001. Four of the five isolates, found in two neighboring regions, appeared to represent a unique clonal group with two subtypes. This clustering of most strains in one PFGE pattern might be explained with the clonal population structure of encapsulated H. influenzae previously observed in Italy (5,9), but further studies are needed to clarify this point. Moreover, since the type e isolates analyzed in this study had no close genetic relationship to the invasive Hib strain circulating in Italy, their derivation from type b isolates by capsular switch is unlikely. H. influenzae PFGE typing has a stronger discriminatory power than biotyping (19). In our study, however, in the isolates assigned to the same clonal group by PFGE, one was classified as biotype I, and the others were biotype IV. These findings suggest that biotyping could be a useful adjunct to PFGE when typing Hie isolates.
In our study, all five Hie isolates contained a single copy of the cap locus, suggesting that they did not possess unusual virulent traits related to the capsule. Since the size of the restriction fragment obtained was smaller than expected on the basis of cap b locus size, a possible explanation may be that the size of region 2 of cap e locus is smaller than that found in Hib strains. Alternatively, the DNA flanking the cap e locus may differ from that found in Hib strains; therefore, the segments upstream and downstream from the cap e region would be smaller.
Both the laboratory-based active surveillance of invasive H. influenzae disease and PCR capsular genotyping could have improved the capability to detect cases attributable to serotypes other than b. Nevertheless, our data suggest the emergence of invasive Hie disease among the adult population in Italy and underline the need to closely monitor infection caused by non–type b strains.
Dr. Cerquetti is a scientist at the Laboratory of Bacteriology and Medical Mycology of Istituto Superiore di Sanità. She has been reference microbiologist for Italy in the European Union Biomedicine and Health II Haemophilus influenzae type b surveillance project (1996–1998) and in the European Union Health and Consumer Protection Directorate-General surveillance network project for invasive H. influenzae disease (2000–2001). Her research interests include investigations on mechanisms of pathogenesis in H. influenzae and Clostridium difficile.
Acknowledgment
We are grateful to Dr. Tonino Sofia for editorial assistance and M.P.E. Slack for providing a prototype type e strain.
References
- Salmaso S, Rota MC. Ciofi degli Atti ML, Tozzi AE, Kreidl P, the ICONA study group. Infant immunization coverage in Italy by cluster survey estimates. Bull World Health Organ. 1999;77:843–51.PubMedGoogle Scholar
- Italian Ministry of Health. Vaccination coverage in Italy. Available from: URL: www.ministerosalute.it
- Squarcione S, Pompa MG, D’Alessandro D. National Surveillance System and Hib meningitis incidence in Italy. Eur J Epidemiol. 1999;15:685–6. DOIPubMedGoogle Scholar
- Ciofi degli Atti ML. Cerquetti M, Tozzi AE, Mastrantonio P, Salmaso S. Haemophilus influenzae invasive disease in Italy, 1997–1998. Eur J Clin Microbiol Infect Dis. 2001;6:436–7.
- Cerquetti M, Ciofi degli Atti ML, Renna G, Tozzi AE, Garlaschi ML, Mastrantonio P, et al. Characterization of non-type b Haemophilus influenzae strains isolated from patients with invasive disease. J Clin Microbiol. 2000;38:4649–52.PubMedGoogle Scholar
- Falla TJ, Crook DWM, Brophy LN, Maskell D, Kroll JS, Moxon ER. PCR for capsular typing of Haemophilus influenzae. J Clin Microbiol. 1994;32:2382–6.PubMedGoogle Scholar
- National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial susceptibility testing; twelfth informational supplement. NCCLS document M100-S12. Vol. 22, No. l. Wayne (PA): The Committee; 2002.
- Corn PG, Anders J, Takala AK, Käyhty H, Hoiseth SK. Genes involved in Haemophilus influenzae type b capsule expression are frequently amplified. J Infect Dis. 1993;167:356–64.PubMedGoogle Scholar
- Tarasi A, D’Ambrosio F, Perrone G, Pantosti A. Susceptibility and genetic relatedness of invasive Haemophilus influenzae type b in Italy. Microb Drug Resist. 1998;4:301–6. DOIPubMedGoogle Scholar
- Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–9.PubMedGoogle Scholar
- Holdaway MD, Turk DC. Capsulated Haemophilus influenzae and respiratory tract disease. Lancet. 1967;1:358–60. DOIPubMedGoogle Scholar
- Talon D, Leroy J, Dupont MJ, Bertrand X, Mermet F, Thouverez M, Antibiotic susceptibility and genotypic characterization of Haemophilus influenzae strains isolated from nasopharyngeal specimens from children in day-care centers in eastern France. Clin Microbiol Infect. 2000;6:519–24. DOIPubMedGoogle Scholar
- Bou R, Dominguez A, Fontanals D, Sanfeliu I, Pons I, Relau J, Prevalence of Haemophilus influenzae pharyngeal carriers in the school population of Catalonia. Eur J Epidemiol. 2000;16:521–6. DOIPubMedGoogle Scholar
- Controni G, Rodriguez WJ, Chang MJ. Meningitis caused by Haemophilus influenzae type e, biotype 4. South Med J. 1982;75:78.PubMedGoogle Scholar
- Waggoner-Fountain LA, Hendley JO, Cody EJ, Perriello VA, Donowitz LG. The emergence of Haemophilus influenzae types e and f as significant pathogens. Clin Infect Dis. 1995;21:1322–4.PubMedGoogle Scholar
- Wu TC, Ferguson RP, Gabel RL. Pneumonia caused by Haemophilus influenzae serotype e. Am J Med Technol. 1982;48:617–9.PubMedGoogle Scholar
- Schlossberg D, Crist AE. Meningitis and septicemia due to Haemophilus influenzae serotype e, biotype IV. Diagn Microbiol Infect Dis. 1985;3:73–5. DOIPubMedGoogle Scholar
- Ogilvie C, Omikunle A, Wang Y, St. Geme JW III, Rodriguez C, Adderson EE. Capsulation loci of non–serotype b encapsulated Haemophilus influenzae. J Infect Dis. 2001;184:144–9. DOIPubMedGoogle Scholar
- Saito M, Umeda A, Yoshida S. Subtyping of Haemophilus influenzae strains by pulsed-field gel electrophoresis. J Clin Microbiol. 1999;37:2142–7.PubMedGoogle Scholar
Figure
Tables
Cite This Article1Members of the Hi Study Group: Istituto Superiore di Sanità, Laboratorio di Epidemiologia e Biostatistica: Patrizia Carbonari; Associazione Microbiologi Clinici Italiani (AMCLI): Pierluigi Nicoletti, Antonio Goglio; Regione Piemonte: Angela Ruggenini Moiraghi, Stefania Orecchia, Annalisa Castella, and Carla Zotti; Provincia Autonoma di Trento: Valter Carraro, Iole Caola, and Anna Calì; Regione Veneto: Giovanni Gallo; Regione Liguria: Pietro Crovari, Cristina Giordano, Pietro Tixi, and Marina Lemmi; Regione Toscana: Paolo Bonanni, Alessia Tomei, Patrizia Pecile, Emanuela Balocchini, and Lucia Pecori; Regione Campania: Francesco Santonastasi, Loredana Cafaro, and Vittorio Pagano; Regione Puglia: Salvatore Barbuti and Maria Chironna.
Table of Contents – Volume 9, Number 2—February 2003
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Marina Cerquetti, Laboratorio di Batteriologia e Micologia Medica, Istituto Superiore di Sanità, Viale Regina Elena, 299, 00161 Roma, Italy; fax: 3906 49387112
Top