Volume 9, Number 5—May 2003
Research
Variant Salmonella Genomic Island 1 Antibiotic Resistance Gene Cluster in Salmonella enterica Serovar Albany
Abstract
Salmonella genomic island 1 (SGI1) contains an antibiotic resistance gene cluster and has been previously identified in multidrug-resistant Salmonella enterica serovars Typhimurium DT104, Agona, and Paratyphi B. We identified a variant SGI1 antibiotic-resistance gene cluster in a multidrug-resistant strain of S. enterica serovar Albany isolated from food fish from Thailand and imported to France. In this strain, the streptomycin resistance aadA2 gene cassette in one of the SGI1 integrons was replaced by a dfrA1 gene cassette, conferring resistance to trimethoprim and an open reading frame of unknown function. Thus, this serovar Albany strain represents the fourth S. enterica serovar in which SGI1 has been identified and the first SGI1 example where gene cassette replacement took place in one of its integron structures. The antibiotic resistance gene cluster of serovar Albany strain 7205.00 constitutes a new SGI1 variant; we propose a name of SGI1-F.
Multidrug-resistant Salmonella enterica serovar Typhimurium definitive phage type 104 (DT104) emerged during the 1980s as a global health problem because of the strain’s involvement in diseases in animals and humans (1). Multidrug-resistant strains of this phage type were first identified from exotic birds in the United Kingdom in the early 1980s and in cattle and humans in the late 1980s; they have since become common in other animal species such as poultry, swine, and sheep. The DT104 epidemic has now spread worldwide, including several outbreaks since 1996 in the United States and Canada (2–5).
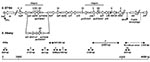
Multidrug-resistant S. enterica serovar Typhimurium DT104 is commonly resistant to ampicillin (Ap), chloramphenicol/florfenicol (Cm/Ff), streptomycin/spectinomycin (Sm/Sp), sulfonamides (Su), and tetracyclines (Tc). The antibiotic resistance genes are clustered in part of a 43-kb genomic island called Salmonella genomic island 1 (SGI1), located between the chromosomal thdf and int2 genes (6,7). The int2 gene is part of a retron that has been detected only in serovar Typhimurium (7). Downstream of the retron sequence is the yidY gene, which is also found in the chromosome of other S. enterica serovars (7). The antibiotic resistance gene cluster represents approximately one third of SGI1 and is located at the 3´ end of the structure (6,7). All resistance genes are clustered and are bracketed by two integron structures (Figure 1) (6–10). The first integron carries the aadA2 gene, which confers resistance to Sm and Sp, and a truncated sul1 (sul1delta) gene. The second integron contains the β-lactamase gene pse-1 conferring resistance to Ap and a complete sul1 gene conferring resistance to Su. Flanked by these two integron structures are the floR gene (8), also called floSt (11) or cmlA-like (9), which confers cross-resistance to Cm and Ff, and the tetracycline-resistance genes tetR and tet(G).
Recently, SGI1 has also been identified in other phage types (i.e., serovar Typhimurium and S. enterica serovars Agona and Paratyphi B), indicating the horizontal transfer potential of SGI1 (6,10,12–15). In serovars Agona and Paratyphi B, SGI1 has the same chromosomal location as in serovar Typhimurium DT104, except that they lack the retron sequence found downstream of SGI1; thus it is located between the thdf gene and the yidY gene of their chromosomes (6,14). Moreover, variant SGI1 antibiotic resistance gene clusters have recently been reported for serovars Typhimurium DT104 and Agona variant SGI1 (12,15). These clusters were probably generated after chromosomal recombinational events, resulting in either deletion or inactivation of some antibiotic resistance genes or in insertion of a new antibiotic resistance gene cassette. In particular, the dfrA10 gene coding for trimethoprim (Tm) resistance was found downstream of the pse-1 integron in two of the SGI1 variant antibiotic resistance gene clusters reported (15). They were accordingly classified in SGI1-A to -E; the resulting antibiotic resistance phenotypes were ApCmFfSmSpSuTcTm, ApSu, SmSpSu, SmSpSuTm, and ApSmSpSuTc, respectively (15).
We examined a strain of S. enterica serovar Albany, isolated from food fish from Thailand and imported in France, that displayed the multidrug-resistance profile ApCmFfSuTcTm. This multidrug resistance profile suggested the possible occurrence of SGI1 with a new variant antibiotic resistance cluster in this serovar.
The S. enterica serovar Albany strain 7205.00 used in this study was isolated from a food fish from Thailand. This strain and control strains S. Typhimurium DT 104 BN9181 (8,13,14), S. Agona 959SA97 (8,13,14), S. Paratyphi B 44 (14), and Escherichia coli strain TOP10 (Invitrogen SARL, Cergy-Pontoise, France) used in cloning experiments were grown at 37°C in brain heart infusion broth or agar plates. The strains were tested for their antibiotic susceptibility by the disc-diffusion assay on Mueller-Hinton plates. Susceptibility was tested by using discs containing the following antibiotics: Ap (10 µg), Cm (30 µg), Ff (30 µg), Sm (10 IU), Sp (100 µg), Su (200 µg), Tc (30 IU), and Tm (5 µg). All antibiotic disks except for Ff were purchased from Bio-Rad (Marnes-la-Coquette, France). Ff disks and the drug itself were obtained from Schering-Plough Animal Health (Kenilworth, NJ). MICs of Ff and Cm were determined by using the standard agar doubling dilution method. MIC breakpoints for Cm and Ff were defined by the Comité de l’Antibiogramme de la Société Française de Microbiologie (CASFM) or by the manufacturer (i.e., susceptible [MIC <8 µg/mL], intermediate [MIC = 16 µg/mL], or resistant [MIC >32 µg/mL]).
Detection of SGI1 and its location were performed by using primers corresponding to left and right (with or without retron) junctions in the chromosome as described (Table; Figure 1) (6,7,14). Polymerase chain reaction (PCR) mapping of the typical antibiotic resistance genes and integrons associated with SGI1 was performed by using conditions and primers as described (Table; Figure 1) (13–15). The antibiotic resistance gene organization was also assessed by Southern blots of genomic DNA cut by either XbaI, XhoI, HindIII or EcoRI by using as a probe the XbaI insert from recombinant plasmid pSTF3, comprising nearly the entire DT104 antibiotic resistance gene cluster, as described (13,14). Presence of SGI1 regions outside the antibiotic resistance gene cluster was assessed by Southern blot of genomic DNA cut by XbaI by using probe p1-9 as described (6,14). This probe contains a 2-kb EcoRI insert corresponding to a central region of SGI1, comprising parts of S023 and S024 open reading frames (ORFs), which code for putative helicase and exonuclease proteins (6).
PCR amplification of the first integron was also performed by using primer int1 of PCR A and primer F3 of PCR B (Figure 1). Cloning of this PCR product in plasmid pCR2.1-TOPO was performed by using the TOPO TA cloning kit (Invitrogen). We used Genome Express (Meylan, France) for nucleotide sequencing of the insert.
Genomic DNA, contained in agarose plugs, was digested with BlnI or XbaI and fragments separated by pulsed-field gel electrophoresis (PFGE) performed by using the CHEF-DR III system (Bio-Rad, Hemel Hempstead, U.K.) at 6 V/cm for 24 h with pulse times of 10–30 sec. The nucleotide sequence of the S. Albany strain 7205.00 integron fragment harboring the Tm resistance gene has been deposited in GenBank under accession number AY146989.
Multidrug-Resistant Phenotype
S. Albany strain 7205.00 showed a multidrug-resistant phenotype similar to SGI1 carrying S. enterica serovars Typhimurium DT104, Agona, or Paratyphi B, (i.e., Ap, Cm/Ff, Su, Tc). The strain was susceptible to Sp and Sm but showed additional resistance to Tm. The strain showed the same level of resistance to Ff as S. enterica serovars Typhimurium DT 104, Agona, or Paratyphi B with a Ff MIC of 64 µg/mL. No plasmids were detected in the S. Albany strain, suggesting a chromosomal location of antibiotic-resistance genes and possibly the presence of SGI1.
Identification of SGI1
To assess the presence of SGI1 and its location in the chromosome of the S. Albany strain 7205.00, PCR was performed by using primers corresponding to the left and right junctions of SGI1 in the Salmonella chromosome (Figure 1). We also used PCR to detect the presence or absence of the int2-retron sequence, which is located downstream of SGI1 in serovar Typhimurium DT104 but not in serovars Agona and Paratyphi B (6,14). PCR results were positive for the left junction of SGI1, as for the other serovars. If a sequence of the int2 gene of the retron was used as reverse primer, the PCR results were negative for the right junction of SGI1 but positive if the sequence of the yidY gene was used. PCR products showed the expected sizes of approximately 500 bp for both the left junction and right junction PCR without the retron sequence, as indicated in Figure 1. These data thus indicate that the serovar Albany strain 7205.00 contains SGI1 at the same chromosomal location as in serovars Typhimurium DT104, Agona, or Paratyphi B (i.e., between the thdf and yidY genes) but lacks the retron sequence found in DT104 and other serovar Typhimurium strains (6,7).
The nucleotide sequences of the left and right junction PCR products were determined by allowing an analysis of the imperfect 18-bp direct repeats flanking SGI1 (6,7). This direct repeat appeared to be a duplication of the last 18 bp of the thdf gene. The right junction direct repeat sequence (DR-R) was previously shown to be identical to the sequence from the respective thdf sequences from sensitive serovar Typhimurium or Agona strains, suggesting the origin of the DR-R is actually the end of thdF (Figure 2) (6). These sequences are slightly divergent between serovars Typhimurium and Agona, with a C located at position 9 of the direct repeat in serovar Typhimurium as opposed to a T at this position in Agona. The left junction direct repeat sequence (DR-L) is identical in both serovars, suggesting the origin of this sequence may be from the donor DNA and not the result of a duplication event. As shown in Figure 2, DR-L and DR-R sequences in the serovar Paratyphi B strain, which were not investigated in a previous study (14), and in the serovar Albany strain 7205.00 are identical to those found in serovar Agona strains carrying SGI1 (Figure 2). These results reinforce the hypothesis that the SGI1 insertions in serovar Typhimurium DT104 and in the other serovars were separate events and not a result of genetic exchange between serovar Typhimurium DT104 and the other serovars.
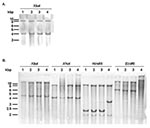
Aside from the antibiotic resistance genes presented below, the presence of the entire SGI1 in the serovar Albany strain 7205.00 was confirmed by Southern blot of XbaI-digested genomic DNA with the p1-9 probe, as described previously. This probe showed XbaI fragments of the expected 4- and 9-kb sizes as in control serovar Typhimurium DT 104, Agona, and Paratyphi B strains carrying SGI1 (Figure 3A).
New Variant Antibiotic Resistance Gene Cluster
PCR mapping of the typical antibiotic resistance genes and integrons associated with SGI1 is schematized in Figure 1. PCR amplifications on genomic DNA extracted from serovar Albany strain 7205.00 yielded fragments B, C, D, E, F, and partial floR of the sizes expected from DNA of serovar Typhimurium DT104 control strain BN9181 (data not shown). However, fragment A specific for the aadA2 integron was not obtained. Thus, these PCR mapping results indicated the presence of floR, tetR, and tet(G) genes and the second integron carrying the pse-1 gene. A positive PCR result for fragment B, representing the link between the aadA2 integron and floR, would nevertheless suggest the presence of at least a partial aadA2 integron. These data are in accordance with the antibiotic resistance phenotype of serovar Albany strain 7205.00 (i.e., ApCmFfSuTc with lack of resistance to Sm and Sp) and indicate the presence of a SGI1 variant antibiotic resistance gene cluster at the level of the aadA2 integron.
These results were confirmed by Southern blot of genomic DNA digested by XbaI, XhoI, HindIII, or EcoRI with the pSTF3 probe containing nearly the entire antibiotic resistance gene cluster of serovar Typhimurium DT104 strain BN9181 (see XbaI fragment in Figure 1). XbaI and XhoI Southern blot profiles of the serovar Albany strain were similar to those obtained for the control strains of serovar Typhimurium DT104, Agona, and Paratyphi B harboring SGI1 (Figure 3B). However, the HindIII and EcoRI Southern blot profiles of the serovar Albany strain were clearly distinct from those of the control strains, confirming the presence of a variant antibiotic resistance gene cluster in the serovar Albany strain.
The genetic variation at the level of the aadA2 integron in the serovar Albany strain was further assessed by PCR with a forward primer of fragment A and reverse primer of fragment B (Figure 1). This PCR result was positive and yielded a fragment approximately 300 bp larger than with DNA from serovar Typhimurium DT104 control strain BN9181 (data not shown). This PCR product was cloned in plasmid pCR2.1-TOPO and sequenced. E. coli carrying this plasmid were resistant to Tm, indicating the presence of a Tm resistance gene in the fragment. Sequence analysis (done by using BLAST [available from: URL: http://www.ncbi.nlm.nih.gov:80/BLAST/]) showed, in addition to the corresponding nucleotide sequence of the DT104 antibiotic resistance gene cluster, two gene cassettes (dfrA1 coding for Tm resistance and an ORF of unknown function), described in class 1 integrons of Vibrio cholerae strains isolated in Thailand and India (99% nucleotide identity; GenBank accession nos. AF221901 and AF455254) (16,17). Thus, instead of the aadA2 gene classically found in the first integron of the SGI1 antibiotic resistance gene cluster, dfrA1 and an ORF of unknown function were found in the corresponding integron of serovar Albany strain 7205.00. The conserved regions of this integron were 100% identical to those found in serovar Typhimurium DT104 with a truncated sul1 gene (Figure 1). The distinct serovar Albany strain 7205.00 HindIII and EcoRI Southern blot profiles with probe pSTF3 described above are in accordance with the nucleotide sequence of the variable region containing dfrA1 of this integron (Figure 1). The antibiotic resistance gene cluster of serovar Albany strain 7205.00 constitutes a new SGI1 variant; we propose a name of SGI1-F, according to previously proposed nomenclature (15).
Evidence for Horizontal Transfer
Macrorestriction analysis by PFGE of the serovar Albany strain DNA cut by XbaI or BlnI showed that the strain is genetically distinct from serovars Typhimurium DT104, Agona, and Paratyphi B in which SGI1 has been identified (Figure 4). This distinction further indicates at the molecular level that the occurrence of SGI1 in the serovar Albany strain probably results from horizontal transfer and not seroconversion of known S. enterica serovars harboring SGI1.
SGI1 is the first genomic island containing an antibiotic resistance gene cluster identified in S. enterica; its acquisition in S. Typhimurium phage type DT104 was possibly an important trait in the worldwide epidemic of the resulting multidrug-resistant clone causing disease in animals as well as in humans. SGI1 has been further identified in other S. enterica serovars, such as in serovar Agona strains isolated from poultry in Belgium and in a serovar Paratyphi B strain isolated from a tropical fish in Singapore (6,13,14). The serovar Albany fish isolate from Thailand in this study represents the fourth S. enterica serovar in which SGI1 has been identified. The identification of SGI1 in several S. enterica serovars, shown by PFGE to be genetically distinct, suggests horizontal transfer of this region. That SGI1 has the same chromosomal location in the different serovars suggests that its insertion occured through site-specific recombination. Genes such as the tandemly arranged int and xis genes found adjacent to the DR-L of SGI1 may play a role in this recombination event because they encode a putative integrase and excisionase, respectively (6). Sequence analysis of the left and right junctions of SGI1 to the Salmonella chromosome provides additional clues about these recombination events in the different serovars. The right junction 18-bp DR-R direct repeat sequence found at the 3´ end of SGI1 appears to be a duplication of the last 18 bp of the thdf gene found upstream of SGI1. The left junction SGI1 18-bp DR-L direct repeat sequence is slightly different from the 3´-end of thdf in sensitive S. enterica serovars lacking SGI1 (Figure 2) but identical in all serovars carrying SGI1. This finding suggests that the origin of this sequence may be from the donor DNA. When SGI1 insertion takes place in a sensitive S. enterica serovar, the last 18 bp of its thdf gene would be duplicated and found at the 3´ end of SGI1 and replaced by the 18 bp of the donor DNA at the 5´ end of SGI1. In other words, the last 18 bp of thdf in sensitive S. enterica serovars may constitute a hotspot for homologous recombination with an 18-bp similar sequence of the SGI1 donor DNA. These DR-R and DR-L minor sequence differences also support the hypothesis that the SGI1 insertions in serovar Typhimurium DT104 and other serovars were separate events and not a result of genetic exchange between serovar Typhimurium DT104 and the other serovars (6). Yet, the origin of SGI1 remains to be determined.
A similar situation has been reported for an approximately 100-kb genomic island in V. cholerae called SXT conjugative, self-transmissible, integrating (constin) element. It carries multiple antibiotic resistance genes, including floR as in SGI1 (18,19). Integration of the element has been experimentally shown to occur through site-specific recombination in a 17-bp sequence found in the circular form of the SXT element and a similar 17-bp sequence of prfC of the V. cholerae and E. coli chromosomes (20). Chromosomal integration and excision of the SXT element required an element-encoded int gene that is found as first gene of the SXT element, as is the int gene of SGI1 (18,20). SGI1 may also exist in an intermediate circular form, but this remains to be demonstrated.
The antibiotic resistance gene cluster of SGI1 contains mainly five antibiotic resistance genes (6–15). Variant SGI1 antibiotic resistance gene clusters have been recently reported in serovars Typhimurium DT104 and Agona containing part of the antibiotic resistance genes or an additional resistance gene (i.e., dfrA10 coding for Tm resistance) (12,15). These variant antibiotic resistance gene clusters were probably generated by recombinational events such as deletions and insertions. The serovar Albany strain of our study represents the first SGI1 example in which gene replacement took place in one of the integron structures. The dfrA1 and ORF gene cassettes found instead of aadA2 may have been introduced by homologous recombination with a class 1 integron containing the same array of gene cassettes from another bacterium (21). Another possibility is the exchange between aadA2 and the two gene cassettes, which would imply excision, mediated by the integron-encoded integrase, of aadA2 and its replacement by the other gene cassettes (22). The array of gene cassettes found in the integron of the serovar Albany strain were the same as those recently reported in integrons of V. cholerae isolated in Thailand and India (16,17). Considering the origin of the serovar Albany strain (i.e., fish exported from Thailand), a possible explanation could be the exchange of antibiotic resistance gene cassettes between epidemic multidrug-resistant V. cholerae strains and Salmonella strains from Thailand. Moreover, during the choleralike epidemic among the Khmers in 1982, children and pregnant women were reportedly treated with trimethoprim-sulfamethoxazole (16,23). A high percentage of Khmer outbreak V. cholerae strains showed resistance to trimethoprim-sulfamethoxazole; the strains that acquired the dfrA1 gene cassette likely became predominant through selective pressure (16,24).
The multidrug-resistant V. cholerae epidemics in humans in Asia might be largely responsible for spread of antibiotic resistance genes. Recent reports describe that human colonization by V. cholerae creates a hyperinfectious bacterial state, which is perpetuated even after purging into natural aquatic reservoirs and may contribute to epidemic spread of cholera (25). These aquatic reservoirs may be an ecologic niche where antibiotic resistance gene exchange takes place between different enterobacterial pathogens. In various seawater places around Hong Kong where untreated sewage is discharged, several enterobacterial pathogens were simultaneously detected such as Salmonella and V. cholerae (26).
As shown in the present study, gene replacement in the integron structures is another way to contribute to variability of the antibiotic resistance gene cluster of SGI1. SGI1 may thus serve as a vehicle of various antibiotic resistance genes in different S. enterica serovars, a situation somewhat similar to that reported for SXT constins and integrons of multidrug-resistant V. cholerae strains (16–20).
Mr. Doublet is a doctoral student at the Institut National de la Recherche Agronomique in France. His main interests are antibiotic resistance mechanisms and the spread of antibiotic-resistance genes in enterobacterial pathogens.
Acknowledgment
We thank C. Mouline for expert technical assistance.
References
- Hancock D, Besser T, Gay J, Rice D, Davis M, Gay C. The global epidemiology of multiresistant Salmonella enterica serovar Typhimurium DT104. In: Brown C, Bolin C, editors. Emerging diseases of animals. Washington: ASM Press; 2000. p. 217–43.
- Besser TE, Goldoft M, Pritchett LC, Khakhria R, Hancock DD, Rice DH, Multiresistant Salmonella Typhimurium DT104 infections of humans and domestic animals in the Pacific Northwest of the United States. Epidemiol Infect. 2000;124:193–200. DOIPubMedGoogle Scholar
- Davis MA, Hancock DD, Besser TE, Rice DH, Gay JM, Gay C, Changes in antimicrobial resistance among Salmonella enterica serovar Typhimurium isolates from humans and cattle in the Northwestern United States, 1982–1997. Emerg Infect Dis. 1999;5:802–6. DOIPubMedGoogle Scholar
- Glynn MK, Bopp C, Dewitt W, Dabney P, Mokhtar M, Angulo FJ. Emergence of multidrug-resistant Salmonella enterica serotype Typhimurium DT104 infections in the United States. N Engl J Med. 1998;338:1333–8. DOIPubMedGoogle Scholar
- Poppe C, Smart N, Khakhria R, Johnson W, Spika J, Prescott J. Salmonella typhimurium DT104: a virulent and drug-resistant pathogen. Can Vet J. 1998;39:559–65.PubMedGoogle Scholar
- Boyd D, Peters GA, Cloeckaert A, Sidi Boumedine K, Chaslus-Dancla E, Imberechts H, Complete nucleotide sequence of a 43-kilobase genomic island associated with the multidrug resistance region of Salmonella enterica serovar Typhimurium DT104 and its identification in phage type DT120 and serovar Agona. J Bacteriol. 2001;183:5725–32. DOIPubMedGoogle Scholar
- Boyd DA, Peters GA, Ng LK, Mulvey MR. Partial characterization of a genomic island associated with the multidrug resistance region of Salmonella enterica Typhimurium DT104. FEMS Microbiol Lett. 2000;189:285–91. DOIPubMedGoogle Scholar
- Arcangioli MA, Leroy-Sétrin S, Martel JL, Chaslus-Dancla E. A new chloramphenicol and florfenicol resistance gene flanked by two integron structures in Salmonella typhimurium DT104. FEMS Microbiol Lett. 1999;174:327–32. DOIPubMedGoogle Scholar
- Briggs CE, Fratamico PM. Molecular characterization of an antibiotic resistance gene cluster of Salmonella typhimurium DT104. Antimicrob Agents Chemother. 1999;43:846–9.PubMedGoogle Scholar
- Cloeckaert A, Schwarz S. Molecular characterization, spread and evolution of multidrug resistance in Salmonella enterica Typhimurium DT104. Vet Res. 2001;32:301–10. DOIPubMedGoogle Scholar
- Bolton LF, Kelley LC, Lee MD, Fedorka-Cray PJ, Maurer JJ. Detection of multi-drug resistant Salmonella enterica serotype typhimurium DT104 based on a gene which confers cross-resistance to florfenicol and chloramphenicol. J Clin Microbiol. 1999;37:1348–51.PubMedGoogle Scholar
- Carattoli A, Filetici E, Villa L, Dionisi AM, Ricci A, Luzzi I. Antibiotic resistance genes and Salmonella genomic island 1 in Salmonella enterica serovar Typhimurium isolated in Italy. Antimicrob Agents Chemother. 2002;46:2821–8. DOIPubMedGoogle Scholar
- Cloeckaert A, Sidi Boumedine K, Flaujac G, Imberechts H, D’Hooghe I, Chaslus-Dancla E. Occurrence of a Salmonella enterica serovar Typhimurium DT104-like antibiotic resistance gene cluster including the floR gene in S. enterica serovar Agona. Antimicrob Agents Chemother. 2000;44:1359–61. DOIPubMedGoogle Scholar
- Meunier D, Boyd D, Mulvey MR, Baucheron S, Mammina C, Nastasi A, Salmonella enterica serotype Typhimurium DT 104 antibiotic resistance genomic island 1 in serotype Paratyphi B. Emerg Infect Dis. 2002;8:430–3. DOIPubMedGoogle Scholar
- Boyd D, Cloeckaert A, Chaslus-Dancla E, Mulvey MR. Characterization of variant Salmonella genomic island 1 multidrug resistance regions from serovars Typhimurium DT104 and Agona. Antimicrob Agents Chemother. 2002;46:1714–22. DOIPubMedGoogle Scholar
- Dalsgaard A, Forslund A, Serichantalergs O, Sandvang D. Distribution and content of class 1 integrons in different Vibrio cholerae O-serotype strains isolated in Thailand. Antimicrob Agents Chemother. 2000;44:1315–21. DOIPubMedGoogle Scholar
- Thungapathra M, Amita, Sinha KK, Ray Chaudhuri S, Garg P, Ramamurthy T, et al. Occurrence of antibiotic resistance gene cassettes aac(6’)-Ib, dfrA5, dfrA12, and ereA2 in class I integrons in non-O1, non-O139 Vibrio cholerae strains in India. Antimicrob Agents Chemother. 2002;46:2948–55. DOIPubMedGoogle Scholar
- Beaber JW, Hochhut B, Waldor MK. Genomic and functional analyses of SXT, an integrating antibiotic resistance gene transfer element derived from Vibrio cholerae. J Bacteriol. 2002;184:4259–69. DOIPubMedGoogle Scholar
- Hochhut B, Lofti Y, Mazel D, Faruque SM, Woodgate R, Waldor MK. Molecular analysis of antibiotic resistance gene clusters in Vibrio cholerae O139 and O1 SXT constins. Antimicrob Agents Chemother. 2001;45:2991–3000. DOIPubMedGoogle Scholar
- Hochhut B, Waldor MK. Site-specific integration of the conjugal Vibrio cholerae SXT element into prfC. Mol Microbiol. 1999;32:99–110. DOIPubMedGoogle Scholar
- Partridge SR, Collis CM, Hall RM. Class 1 integron containing a new gene cassette, aadA10, associated with Tn1404 from R151. Antimicrob Agents Chemother. 2002;46:2400–8. DOIPubMedGoogle Scholar
- Hall RM, Collis CM. Mobile gene cassettes and integrons: capture and spread of genes by site-specific recombination. Mol Microbiol. 1995;15:593–600. DOIPubMedGoogle Scholar
- Bagchi K, Echeverria P, Arthur JD, Sethabutr O, Serichantalergs O, Hoge CW. Epidemic of diarrhea caused by Vibrio cholerae non-O1 that produced heat-stable toxin among Khmers in a camp in Thailand. J Clin Microbiol. 1993;31:1315–7.PubMedGoogle Scholar
- Dalsgaard A, Serichantalergs O, Pitarangsi C, Echeverria P. Molecular characterization and antibiotic susceptibility patterns of Vibrio cholerae non-O1. Epidemiol Infect. 1995;114:51–63. DOIPubMedGoogle Scholar
- Merrell DS, Butler SM, Qadri F, Dolganov NA, Alam A, Cohen MB, Host-induced epidemic spread of the cholera bacterium. Nature. 2002;417:642–5. DOIPubMedGoogle Scholar
- Kong RYC, Lee SKY, Law TWF, Law SHW, Wu RSS. Rapid detection of six types of bacterial pathogens in marine waters by multiplex PCR. Water Res. 2002;36:2802–12. DOIPubMedGoogle Scholar
Figures
Table
Cite This ArticleTable of Contents – Volume 9, Number 5—May 2003
| EID Search Options |
|---|
|
|
|
|
|
|


Please use the form below to submit correspondence to the authors or contact them at the following address:
Axel Cloeckaert, Unité BioAgresseurs, Santé, Environnement, Institut National de la Recherche Agronomique, 37380 Nouzilly, France; fax: (33) 2 47 42 77 74
Top