Volume 9, Number 3—March 2003
Dispatch
Rabies in Sri Lanka: Splendid Isolation
Figure 1
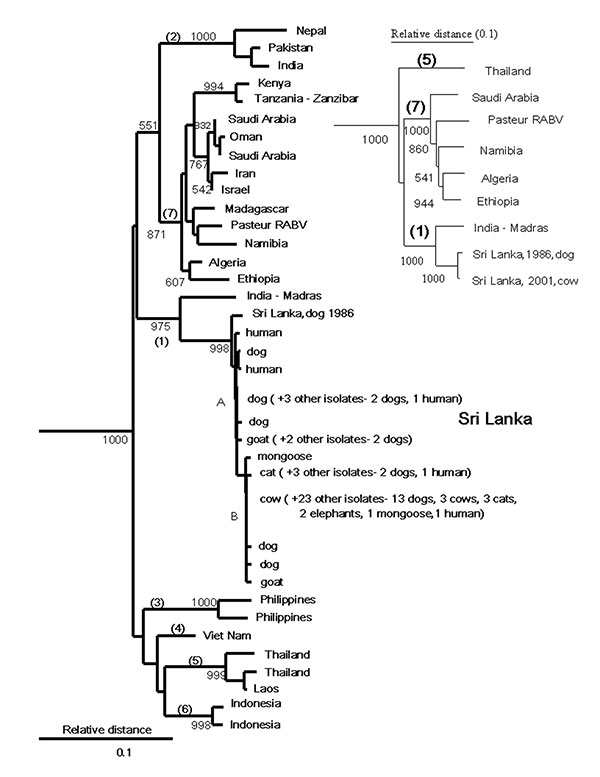
Figure 1. Neighbor-joining tree for 264 base pairs (bp) of nucleoprotein gene sequence (bp 1157–1420) for 44 rabies virus samples from Sri Lanka compared with samples from other areas of Asia and Africa with uncontrolled dog rabies. Samples were aligned with Pasteur rabies virus (GenBank accession no. M13215) by using the PileUp program of the Wisconsin Package version 10 (Genetic Computer Group, 1999, Madison, WI). The phylogenetic analyses were perfomed by using the DNADIST (Kimura two-parameter method), Neighbor (Neighbor-joining method), SEQBOOT, and CONSENSE programs of the PHYLIP package, version 3.5 (available from: URL: http://evolution.genetics.washington.edu/phylip.html). The program TREEVIEW was used to obtain the graphic output (available from: URL: http://taxonomy.zoology.gla.ac.uk/rod/treeview.html). Australian bat lyssavirus (GenBank AF081020) and Duvenhage virus (GenBank U22848) were used as outgroups (not shown). Bootstrap values >50% in 1,000 resamplings of the data are shown at nodes corresponding to the different lineages and sample clusters. The nodes signifying highest order clustering of rabies virus lineages (supported by bootstrap values >70%) are numbered 1–7. The two clusters of Sri Lankan samples, designated by mutations at nucleotides 1231 and 1408, are indicated as A and B. Inset shows a similar tree achieved by analysis of complete nucleoprotein gene sequence (1414 bp). All nonidentical Sri Lankan samples were submitted to GenBank (AY138549–AY138564). The Table lists sample collection information.