Volume 24, Number 3—March 2018
Research
Capsule Typing of Haemophilus influenzae by Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry1
Figure 1
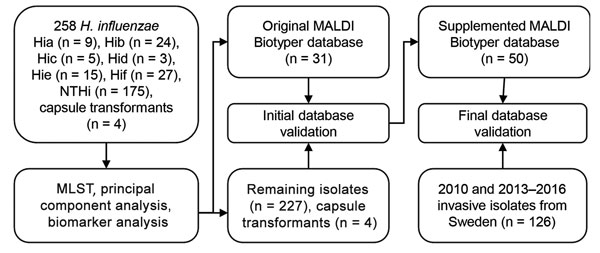
Figure 1. Culture collections and methods used in this study for capsule typing of Haemophilus influenzae by MALDI-TOF mass spectrometry. An evaluation set of H. influenzae isolates of all capsule types from diverse geographic origins and time periods and isogenic capsule transformants (30) were used to investigate capsule type-specific differences in MALDI-TOF mass spectra. MLST was used to ensure adequate coverage of different genetic lineages of encapsulated H. influenzae. Reference isolates from the evaluation set (encapsulated and nonencapsulated) were selected to construct a new typing database in MALDI Biotyper. This database was tested with the remaining isolates in the set, and misclassified isolates were added to the database. The final supplemented database was blindly validated with a second culture collection that consisted of clinical invasive isolates. Hia, H. influenzae type a; Hib, H. influenzae type b; Hic, H. influenzae type c; Hid, H. influenzae type d; Hie, H. influenzae type e; Hif, H. influenzae type f; MALDI-TOF, matrix-assisted laser desorption/ionization time-of-flight; MLST, multilocus sequence typing; NTHi, nontypeable H. influenzae.
References
- Pittman M. Variation and type specificity in the bacterial species Haemophilus influenzae. J Exp Med. 1931;53:471–92. DOIPubMedGoogle Scholar
- Peltola H. Worldwide Haemophilus influenzae type b disease at the beginning of the 21st century: global analysis of the disease burden 25 years after the use of the polysaccharide vaccine and a decade after the advent of conjugates. Clin Microbiol Rev. 2000;13:302–17. DOIPubMedGoogle Scholar
- Langereis JD, de Jonge MI. Invasive disease caused by nontypeable Haemophilus influenzae. Emerg Infect Dis. 2015;21:1711–8. DOIPubMedGoogle Scholar
- Resman F, Ristovski M, Ahl J, Forsgren A, Gilsdorf JR, Jasir A, et al. Invasive disease caused by Haemophilus influenzae in Sweden 1997-2009; evidence of increasing incidence and clinical burden of non-type b strains. Clin Microbiol Infect. 2011;17:1638–45. DOIPubMedGoogle Scholar
- Ladhani S, Slack MP, Heath PT, von Gottberg A, Chandra M, Ramsay ME; European Union Invasive Bacterial Infection Surveillance participants. Invasive Haemophilus influenzae Disease, Europe, 1996-2006. Emerg Infect Dis. 2010;16:455–63. DOIPubMedGoogle Scholar
- Whittaker R, Economopoulou A, Dias JG, Bancroft E, Ramliden M, Celentano LP; European Centre for Disease Prevention and Control Country Experts for Invasive Haemophilus influenzae Disease. Epidemiology of Invasive Haemophilus influenzae Disease, Europe, 2007-2014. Emerg Infect Dis. 2017;23:396–404. DOIPubMedGoogle Scholar
- Bender JM, Cox CM, Mottice S, She RC, Korgenski K, Daly JA, et al. Invasive Haemophilus influenzae disease in Utah children: an 11-year population-based study in the era of conjugate vaccine. Clin Infect Dis. 2010;50:e41–6. DOIPubMedGoogle Scholar
- Bruce MG, Zulz T, DeByle C, Singleton R, Hurlburt D, Bruden D, et al. Haemophilus influenzae serotype a invasive disease, Alaska, USA, 1983-2011. Emerg Infect Dis. 2013;19:932–7. DOIPubMedGoogle Scholar
- Zanella RC, Bokermann S, Andrade AL, Flannery B, Brandileone MC. Changes in serotype distribution of Haemophilus influenzae meningitis isolates identified through laboratory-based surveillance following routine childhood vaccination against H. influenzae type b in Brazil. Vaccine. 2011;29:8937–42. DOIPubMedGoogle Scholar
- Rotondo JL, Sherrard L, Helferty M, Tsang R, Desai S. The epidemiology of invasive disease due to Haemophilus influenzae serotype a in the Canadian North from 2000 to 2010. Int J Circumpolar Health. 2013;72:72. DOIPubMedGoogle Scholar
- Bajanca-Lavado MP, Simões AS, Betencourt CR, Sá-Leão R; Portuguese Group for Study of Haemophilus influenzae invasive infection. Characteristics of Haemophilus influenzae invasive isolates from Portugal following routine childhood vaccination against H. influenzae serotype b (2002-2010). Eur J Clin Microbiol Infect Dis. 2014;33:603–10. DOIPubMedGoogle Scholar
- Ladhani SN, Collins S, Vickers A, Litt DJ, Crawford C, Ramsay ME, et al. Invasive Haemophilus influenzae serotype e and f disease, England and Wales. Emerg Infect Dis. 2012;18:725–32. DOIPubMedGoogle Scholar
- Ladhani S, Heath PT, Slack MP, McIntyre PB, Diez-Domingo J, Campos J, et al.; Participants of the European Union Invasive Bacterial Infections Surveillance Network. Haemophilus influenzae serotype b conjugate vaccine failure in twelve countries with established national childhood immunization programmes. Clin Microbiol Infect. 2010;16:948–54. DOIPubMedGoogle Scholar
- Ladhani SN. Two decades of experience with the Haemophilus influenzae serotype b conjugate vaccine in the United Kingdom. Clin Ther. 2012;34:385–99. DOIPubMedGoogle Scholar
- Juarez MD, Rancaño C, Neyro S, Biscayart C, Katz N, Pasinovich M, et al. What’s happening with Haemophilus influenzae type B invasive disease in Latin America region? Argentina’s experience. In: Abstracts of IDWeek 2016, New Orleans, October 26–30. Abstract no. 768 [cited 2017 Dec 18]. https://idsa.confex.com/idsa/2016/webprogram/POSTER.html
- World Health Organization. Global and regional immunization profile, 2016 global summary [cited 2017 Nov 9]. http://www.who.int/immunization/monitoring_surveillance/data/gs_gloprofile.pdf?ua=1
- Musser JM, Kroll JS, Moxon ER, Selander RK. Clonal population structure of encapsulated Haemophilus influenzae. Infect Immun. 1988;56:1837–45.PubMedGoogle Scholar
- Meats E, Feil EJ, Stringer S, Cody AJ, Goldstein R, Kroll JS, et al. Characterization of encapsulated and noncapsulated Haemophilus influenzae and determination of phylogenetic relationships by multilocus sequence typing. J Clin Microbiol. 2003;41:1623–36. DOIPubMedGoogle Scholar
- Erwin AL, Sandstedt SA, Bonthuis PJ, Geelhood JL, Nelson KL, Unrath WC, et al. Analysis of genetic relatedness of Haemophilus influenzae isolates by multilocus sequence typing. J Bacteriol. 2008;190:1473–83. DOIPubMedGoogle Scholar
- University of Oxford. Haemophilus influenzae MLST website [cited 2017 Nov 9]. http://pubmlst.org/hinfluenzae/
- Myers AL, Jackson MA, Zhang L, Swanson DS, Gilsdorf JR. Haemophilus influenzae type b invasive disease in Amish children, Missouri, USA, 2014. Emerg Infect Dis. 2017;23:112–4. DOIPubMedGoogle Scholar
- LaClaire LL, Tondella ML, Beall DS, Noble CA, Raghunathan PL, Rosenstein NE, et al.; Active Bacterial Core Surveillance Team Members. Identification of Haemophilus influenzae serotypes by standard slide agglutination serotyping and PCR-based capsule typing. J Clin Microbiol. 2003;41:393–6. DOIPubMedGoogle Scholar
- Satola SW, Collins JT, Napier R, Farley MM. Capsule gene analysis of invasive Haemophilus influenzae: accuracy of serotyping and prevalence of IS1016 among nontypeable isolates. J Clin Microbiol. 2007;45:3230–8. DOIPubMedGoogle Scholar
- Falla TJ, Crook DW, Brophy LN, Maskell D, Kroll JS, Moxon ER. PCR for capsular typing of Haemophilus influenzae. J Clin Microbiol. 1994;32:2382–6.PubMedGoogle Scholar
- Davis GS, Sandstedt SA, Patel M, Marrs CF, Gilsdorf JR. Use of bexB to detect the capsule locus in Haemophilus influenzae. J Clin Microbiol. 2011;49:2594–601. DOIPubMedGoogle Scholar
- Lâm TT, Elias J, Frosch M, Vogel U, Claus H. New diagnostic PCR for Haemophilus influenzae serotype e based on the cap locus of strain ATCC 8142. Int J Med Microbiol. 2011;301:176–9. DOIPubMedGoogle Scholar
- van Ketel RJ, de Wever B, van Alphen L. Detection of Haemophilus influenzae in cerebrospinal fluids by polymerase chain reaction DNA amplification. J Med Microbiol. 1990;33:271–6. DOIPubMedGoogle Scholar
- Clark AE, Kaleta EJ, Arora A, Wolk DM. Matrix-assisted laser desorption ionization-time of flight mass spectrometry: a fundamental shift in the routine practice of clinical microbiology. Clin Microbiol Rev. 2013;26:547–603. DOIPubMedGoogle Scholar
- Månsson V, Resman F, Kostrzewa M, Nilson B, Riesbeck K. Identification of Haemophilus influenzae type b isolates by use of matrix-assisted laser desorption ionization−time of flight mass spectrometry. J Clin Microbiol. 2015;53:2215–24. DOIPubMedGoogle Scholar
- Zwahlen A, Kroll JS, Rubin LG, Moxon ER. The molecular basis of pathogenicity in Haemophilus influenzae: comparative virulence of genetically-related capsular transformants and correlation with changes at the capsulation locus cap. Microb Pathog. 1989;7:225–35. DOIPubMedGoogle Scholar
- Alexander HE, Leidy G. Determination of inherited traits of H. influenzae by desoxyribonucleic acid fractions isolated from type-specific cells. J Exp Med. 1951;93:345–59. DOIPubMedGoogle Scholar
- Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 2012;9:772. DOIPubMedGoogle Scholar
- Guindon S, Gascuel O, Rannala B. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. DOIPubMedGoogle Scholar
- López-Fernández H, Santos HM, Capelo JL, Fdez-Riverola F, Glez-Peña D, Reboiro-Jato M. Mass-Up: an all-in-one open software application for MALDI-TOF mass spectrometry knowledge discovery. BMC Bioinformatics. 2015;16:318. DOIPubMedGoogle Scholar
- Kehrmann J, Wessel S, Murali R, Hampel A, Bange FC, Buer J, et al. Principal component analysis of MALDI TOF MS mass spectra separates M. abscessus (sensu stricto) from M. massiliense isolates. BMC Microbiol. 2016;16:24. DOIPubMedGoogle Scholar
- Rizzardi K, Åkerlund T. High molecular weight typing with MALDI-TOF MS: a novel method for rapid typing of Clostridium difficile. PLoS One. 2015;10:e0122457. DOIPubMedGoogle Scholar
- Camoez M, Sierra JM, Dominguez MA, Ferrer-Navarro M, Vila J, Roca I. Automated categorization of methicillin-resistant Staphylococcus aureus clinical isolates into different clonal complexes by MALDI-TOF mass spectrometry. Clin Microbiol Infect. 2016;22:161.e1–7. DOIPubMedGoogle Scholar
- Josten M, Reif M, Szekat C, Al-Sabti N, Roemer T, Sparbier K, et al. Analysis of the matrix-assisted laser desorption ionization-time of flight mass spectrum of Staphylococcus aureus identifies mutations that allow differentiation of the main clonal lineages. J Clin Microbiol. 2013;51:1809–17. DOIPubMedGoogle Scholar
- Ojima-Kato T, Yamamoto N, Suzuki M, Fukunaga T, Tamura H. Discrimination of Escherichia coli O157, O26 and O111 from other serovars by MALDI-TOF MS based on the S10-GERMS method. PLoS One. 2014;9:e113458. DOIPubMedGoogle Scholar
- Fothergill LD, Chandler CA, Spencer M. Observations on the dissociation of meningitic strains of H. influenzae. J Immunol. 1936;31:401–15.
- Hoiseth SK, Connelly CJ, Moxon ER. Genetics of spontaneous, high-frequency loss of b capsule expression in Haemophilus influenzae. Infect Immun. 1985;49:389–95.PubMedGoogle Scholar
- Tsang RS, Li YA, Mullen A, Baikie M, Whyte K, Shuel M, et al. Laboratory characterization of invasive Haemophilus influenzae isolates from Nunavut, Canada, 2000-2012. Int J Circumpolar Health. 2016;75:29798. DOIPubMedGoogle Scholar
- Sauget M, Valot B, Bertrand X, Hocquet D. Can MALDI-TOF mass spectrometry reasonably type bacteria? Trends Microbiol. 2017;25:447–55. DOIPubMedGoogle Scholar
1Preliminary results from this study were presented at the IDWeek 2016 Conference, October 26−30, 2016, New Orleans, Louisiana, USA.
2These senior authors contributed equally to this article.