Volume 28, Number 5—May 2022
Dispatch
Lack of Evidence for Crimean–Congo Hemorrhagic Fever Virus in Ticks Collected from Animals, Corsica, France
Abstract
In Corsica, France, 9.1% of livestock serum samples collected during 2014–2016 were found to have antibodies against Crimean–Congo hemorrhagic fever virus (CCHFV), an emerging tickborne zoonotic disease. We tested 8,051 ticks for CCHFV RNA and Nairovirus RNA. The results indicate that Corsica is not a hotspot for CCHFV.
Crimean–Congo hemorrhagic fever (CCHF) is a tickborne zoonotic disease that is characterized by hemorrhagic fever and can progress from mild, nonspecific signs to a severe and fatal hemorrhagic disease. The CCHF virus (CCHFV) is an enveloped, segmented, negative-sense, single-stranded RNA member of the family Nairoviridae, genus Orthonairovirus. CCHFV has been detected in >35 species of ticks worldwide, among which ticks belonging to the genus Hyalomma are the primary vectors in humans and wild and domestic animals (1). Humans are infected through tick bites and direct contact with infected blood and body fluids during occupational exposure (e.g., farming, slaughtering, and medical and nursing care).
CCHF is endemic in Africa, Asia, and the Balkan region (2). In Western Europe, autochthonous human cases were reported only in Spain, where CCHFV was identified in H. lusitanicum ticks (3). In Corsica, a French Mediterranean island, 9.1% of livestock (i.e., cattle, goats, sheep) serum samples contained CCHFV-specific IgG during 2014–2016 (4). Entomologic surveys revealed that the H. marginatum tick, a vector of CCHFV, was present in Corsica (5).
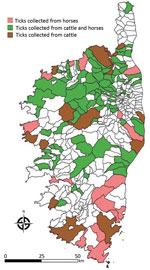
To assess whether CCHFV circulates in Corsica, we collected 8,051 ticks from wild and domestic animals in selected sites on the island during 2016–2020 (Table, Figure). These 8,051 ticks included 7,156 ticks taken from 3,674 domestic animals and 895 ticks taken from 188 wild animals. They consisted of 4,177 Rhipicephalus bursa (51.8%), 2,386 H. marginatum (29.6%), 839 Dermacentor marginatus (10.4%) and 282 H. scupense (3.5%) ticks. We identified ticks at the species level by using a pictorial guide and confirmed morphologic identification by using sequencing of mitochondrial 16S rDNA (5). We then pooled up to 10 ticks per pool on the basis of developmental stage (nymphs, nonengorged females, and male adults) and host (Table). Pools, containing an average of 2.5 ticks (range 1–10 ticks) were crushed in phosphate-buffered saline with TissueLyser II (QIAGEN, https://www.qiagen.com) at 5,500 rpm for 3 min. We spiked each pool before extraction with a predefined amount of MS2 bacteriophage to monitor the subsequent steps (nucleic acid extraction, reverse transcription, and PCR amplification) and to detect the presence of inhibitors and enzymatic reactions as described (6). We performed DNA extraction by using QIAcube HT and a QIAamp cador Pathogen Mini Kit (QIAGEN), according to the manufacturer’s instructions. We eluted DNA in 150 μL of buffer and stored at –20°C. We tested each pool for the presence of CCHFV RNA by using a real-time, reverse transcription PCR (7) and the presence of Nairovirus RNA by using a pangeneric reverse transcription PCR (8).
We detected neither CCHFV RNA nor Nairovirus RNA in the 8,051 ticks. The absence of CCHFV or Nairovirus RNA was not attributable to technical problems or presence of inhibitors, which were ruled out by MS2 bacteriophage monitoring. Moreover, we detected viral RNA corresponding to new tickborne Phleboviruses in 40 samples (5%) and Flavivirus in 7 samples (0.9%); these samples remain under investigation, and results will be reported after detailed characterization.
We considered whether CCHFV RNA was not detected because of a low minimum infection rate (MIR) that a larger number of ticks would have been required. We calculated the theoretical power that could be achieved by using the number of ticks obtained in our study. On the basis of an expected CCHFV prevalence (P) of ≈0.2% and a pool size (k) of 2 ticks, a total of 7,676 ticks have to be tested for a prevalence estimation with a 95% CI and a precision (d) set at +0.001 because the disease prevalence is <0.1 (10%) (9). Thus, with a sample of 8,051 ticks, we were able to detect a prevalence of >0.2%.
The rate of CCHFV-infected ticks in countries in Europe with enzootic foci ranges from 0.50% to 3.70% among Hyalomma spp. ticks (2.8% [44/1,579 H. lusitanicum ticks] in Spain, 3.7% [6/161 H. marginatum ticks] in Bulgaria, and 0.5% [1/199 H. marginatum ticks] in Kosovo) and from 1.5% to 6.2% among Rhipicephalus spp. (1.5% [2/123 R. sanguineus ticks] in Bulgaria and 6.2% (8/130 R. bursa ticks] in Kosovo) (10). Other studies conducted outside of Europe have largely reported MIR values >0.2% among ticks: 0.71% in South Africa (1.6% [15/914] H. truncatum and 0.2% [2/1,149] H. rufipes) (11); 2.6% in Mauritania (39/1,517 Hyalomma spp.) (12); 3.8% (20/525 Hyalomma spp.) in Pakistan (13); and 51.5% (103/200 H. marginatum) in Turkey (14). These studies were conducted during the past 5 years using methods comparable to those of our study. The number of Hyalomma (n = 2,682) and Rhipicephalus (n = 4,177) ticks that we tested are much higher than reported in these previous studies. Therefore, our study would have been able to recognize CCHFV presence for a prevalence >0.2%, which is 10 times lower than the lowest overall prevalence value reported to date in countries where CCHFV is present: 2.1% (95% CI 1.3%–2.9%) according to a recent meta-analysis (10). Furthermore, another study addressing the presence of CCHFV RNA in Hyalomma spp. ticks (362 H. marginatum and 135 H. scupense) and Rhipicephalus ticks (n = 518) collected in 2014 from domestic and wild animals in Corsica also provided only negative results (15). In all countries where CCHF cases are described, the observed MIR of ticks is >2.5 times higher than the detection limit in our study (0.2%). Another argument that strongly supports the contention that the lack of detection of CCHFV or Nairovirus RNA was not caused by technical problems is based on the consideration that the protocol used in this study enables the detection of a wide variety of different CCHFV strains, a fact that confirms the accuracy of the results (7,8).
Recent studies to determine whether CCHFV is present in Corsica and to what extent it is a threat for human populations provide contrasting data. On one hand, tick species that are able to transmit CCHFV are present and widely distributed, and a serologic study based on ELISA screening and neutralization test for confirmation supports the presence of CCHFV or an antigenically related agent. On the other hand, the absence of detection of CCHFV RNA (or an antigenically related agent) in a large number of ticks, together with the absence of a CCHF case, supports the absence of CCHFV in Corsica to date.
In any case, the absence of a documented case of CCHF together with the lack of detection of CCHFV RNA in tick species that are recognized as a competent vector enables us to declare that Corsica is not a hotspot for CCHFV and that the threat to the human population is very limited. However, this discrepant set of data pleads for a One Health approach for dealing with the CCHF question in Corsica and the potential exposure of island population. To do so, the roadmap established by the World Health Organization’s R&D blueprint (https://www.who.int/teams/blueprint/about) should be followed. Because the accuracy of CCHFV serologic assays has been questioned, several tests must be combined as advocated. Then, serologic studies in animals and humans must be synchronized with virus detection in ticks and systematic screening of patients with uncharacterized febrile illness during the tick season. A need exists for a large-scale One Health prospective program for surveillance of ticks, vertebrates, and humans in Corsica.
Mr. Cicculli is a doctoral student at the University of Corsica and Aix-Marseille University. His research interests include the epidemiology of vectorborne pathogens.
Acknowledgments
We are grateful to the staff of the slaughterhouse at Ponte-Leccia for their help in collecting ticks from cattle.
This study has been funded in part by the European Virus Archive Global Project, which has received funding from the European Union’s Horizon 2020-INFRAIA-2019 research and innovation programme (grant agreement no. 871029) and by the European Union–Cepheid Innovative Medicines Initiative Joint Undertaking (as part of the Viral Haemorrhagic Fever: Modern Approaches For Developing Bedside Rapid Diagnostics [VHFMoDRAD] under grant no. 823666).
References
- Spengler JR, Bergeron É, Rollin PE. Seroepidemiological studies of Crimean-Congo hemorrhagic fever virus in domestic and wild animals. PLoS Negl Trop Dis. 2016;10:
e0004210 . DOIPubMedGoogle Scholar - Portillo A, Palomar AM, Santibáñez P, Oteo JA. Epidemiological aspects of Crimean-Congo hemorrhagic fever in Western Europe: what about the future? Microorganisms. 2021;9:649. DOIPubMedGoogle Scholar
- Negredo A, Sánchez-Ledesma M, Llorente F, Pérez-Olmeda M, Belhassen-García M, González-Calle D, et al. Retrospective identification of early autochthonous case of Crimean-Congo hemorrhagic fever, Spain, 2013. Emerg Infect Dis. 2021;27:1754–6. DOIPubMedGoogle Scholar
- Grech-Angelini S, Lancelot R, Ferraris O, Peyrefitte CN, Vachiery N, Pédarrieu A, et al. Crimean-Congo hemorrhagic fever virus antibodies among livestock on Corsica, France, 2014–2016. Emerg Infect Dis. 2020;26:1041–4. DOIPubMedGoogle Scholar
- Cicculli V, Oscar M, Casabianca F, Villechenaud N, Charrel R, de Lamballerie X, et al. Molecular Detection of Spotted-Fever Group Rickettsiae in ticks collected from domestic and wild animals in Corsica, France. Pathogens. 2019;8:138. DOIPubMedGoogle Scholar
- Ninove L, Nougairede A, Gazin C, Thirion L, Delogu I, Zandotti C, et al. RNA and DNA bacteriophages as molecular diagnosis controls in clinical virology: a comprehensive study of more than 45,000 routine PCR tests. PLoS One. 2011;6:
e16142 . DOIPubMedGoogle Scholar - Wölfel R, Paweska JT, Petersen N, Grobbelaar AA, Leman PA, Hewson R, et al. Virus detection and monitoring of viral load in Crimean-Congo hemorrhagic fever virus patients. Emerg Infect Dis. 2007;13:1097–100. DOIPubMedGoogle Scholar
- Lambert AJ, Lanciotti RS. Consensus amplification and novel multiplex sequencing method for S segment species identification of 47 viruses of the Orthobunyavirus, Phlebovirus, and Nairovirus genera of the family Bunyaviridae. J Clin Microbiol. 2009;47:2398–404. DOIPubMedGoogle Scholar
- Ausvet. Epitools—epidemiological calculators. 2016 [cited 2022 Feb 19]. https://epitools.ausvet.com.au
- Belobo JTE, Kenmoe S, Kengne-Nde C, Emoh CPD, Bowo-Ngandji A, Tchatchouang S, et al. Worldwide epidemiology of Crimean-Congo Hemorrhagic Fever Virus in humans, ticks and other animal species, a systematic review and meta-analysis. PLoS Negl Trop Dis. 2021;15:
e0009299 . DOIPubMedGoogle Scholar - Msimang V, Weyer J, le Roux C, Kemp A, Burt FJ, Tempia S, et al. Risk factors associated with exposure to Crimean-Congo haemorrhagic fever virus in animal workers and cattle, and molecular detection in ticks, South Africa. PLoS Negl Trop Dis. 2021;15:
e0009384 . DOIPubMedGoogle Scholar - Schulz A, Barry Y, Stoek F, Pickin MJ, Ba A, Chitimia-Dobler L, et al. Detection of Crimean-Congo hemorrhagic fever virus in blood-fed Hyalomma ticks collected from Mauritanian livestock. Parasit Vectors. 2021;14:342. DOIPubMedGoogle Scholar
- Kasi KK, von Arnim F, Schulz A, Rehman A, Chudhary A, Oneeb M, et al. Crimean-Congo haemorrhagic fever virus in ticks collected from livestock in Balochistan, Pakistan. Transbound Emerg Dis. 2020;67:1543–52. DOIPubMedGoogle Scholar
- Akyildiz G, Bente D, Keles AG, Vatansever Z, Kar S. High prevalence and different genotypes of Crimean-Congo hemorrhagic fever virus genome in questing unfed adult Hyalomma marginatum in Thrace, Turkey. Ticks Tick Borne Dis. 2021;12:
101622 . DOIPubMedGoogle Scholar - Grech-Angelini S, Stachurski F, Vayssier-Taussat M, Devillers E, Casabianca F, Lancelot R, et al. Tick-borne pathogens in ticks (Acari: Ixodidae) collected from various domestic and wild hosts in Corsica (France), a Mediterranean island environment. Transbound Emerg Dis. 2020;67:745–57. DOIPubMedGoogle Scholar
Figure
Table
Cite This ArticleOriginal Publication Date: April 06, 2022
Table of Contents – Volume 28, Number 5—May 2022
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Vincent Cicculli, Laboratoire de virologie, UR7310 Université de Corse, 20250, Corte, France
Top