Volume 30, Number 8—August 2024
Dispatch
Macrolide-Resistant Mycoplasma pneumoniae Infections among Children before and during COVID-19 Pandemic, Taiwan, 2017–2023
Figure 2
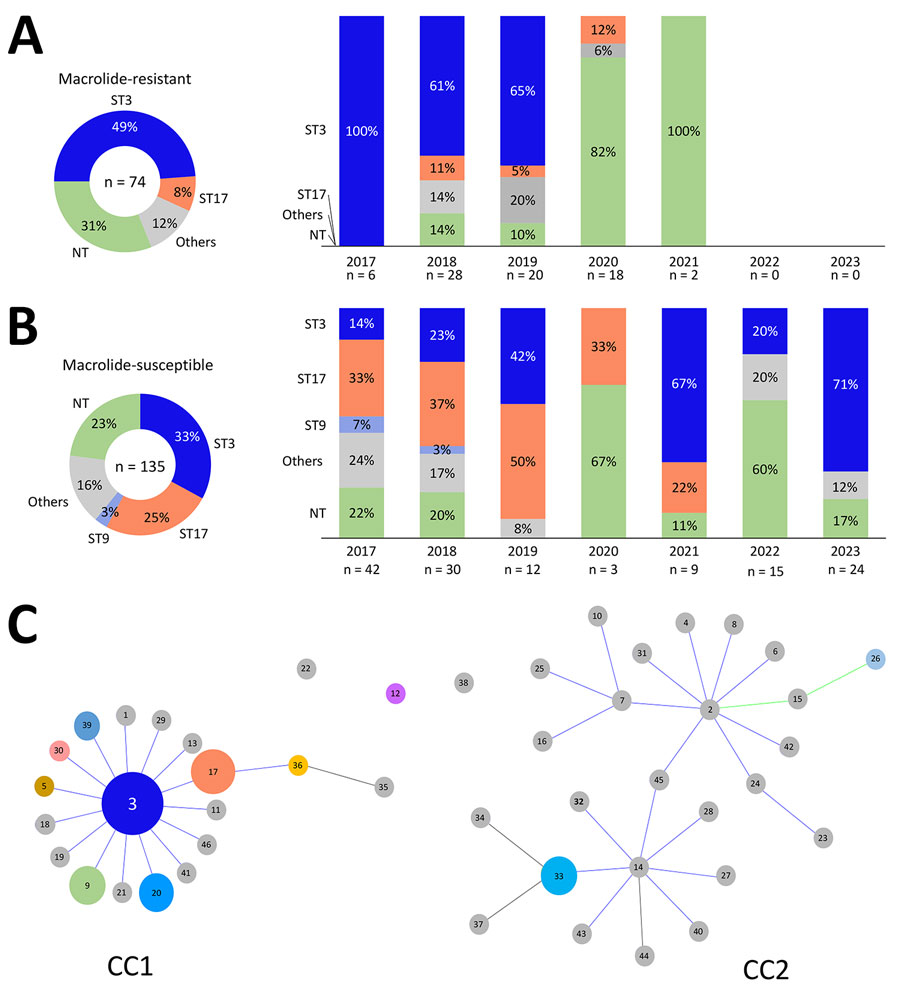
Figure 2. Relationships between year of isolation, ST, and genotype distribution in a study of macrolide-resistant Mycoplasma pneumoniae infections among children before and during the COVID-19 pandemic, Taiwan, 2017–2023. A) Macrolide-resistant M. pneumoniae isolates (n = 74) and 5 identified STs: ST3, ST17, ST26, ST33, and ST36. Macrolide resistance substantially decreased after 2021. ST3 was the predominant strain in macrolide-resistant isolates, especially during 2017–2019. B) Macrolide-susceptible M. pneumoniae isolates (n = 135) and 10 identified STs: ST3, ST17, ST9, ST5, ST12, ST20, ST30, ST33, ST39, and ST46. ST17 was the predominant strain during 2017–2019. ST3 was the most common strain and was distributed across all years. C) The relationship between M. pneumoniae CC and ST depicted by goeBURST (https://phyloviz.readthedocs.io/en/latest/data_analysis.html). The data, comprising 144 strains from Taiwan (2017–2023) and previously reported STs (shown in gray) from PubMLST (https://pubmlst.org), demonstrate the genetic relationships within the dataset analyzed by the goeBURST algorithm. The size of each circle is proportional to the number of isolates for each ST, and most STs belonged to CC1, including the leading 2 STs, ST3, and ST17. Green sections indicate M. pneumoniae strains could not be successfully identified using multilocus sequence typing. CC, clonal complex; NT, nontypable; ST, sequence type.