Disclaimer: Early release articles are not considered as final versions. Any changes will be reflected in the online version in the month the article is officially released.
Volume 31, Number 5—May 2025
Research Letter
Autochthonous Leishmania (Viannia) lainsoni in Dog, Rio de Janeiro State, Brazil, 2023
Figure 2
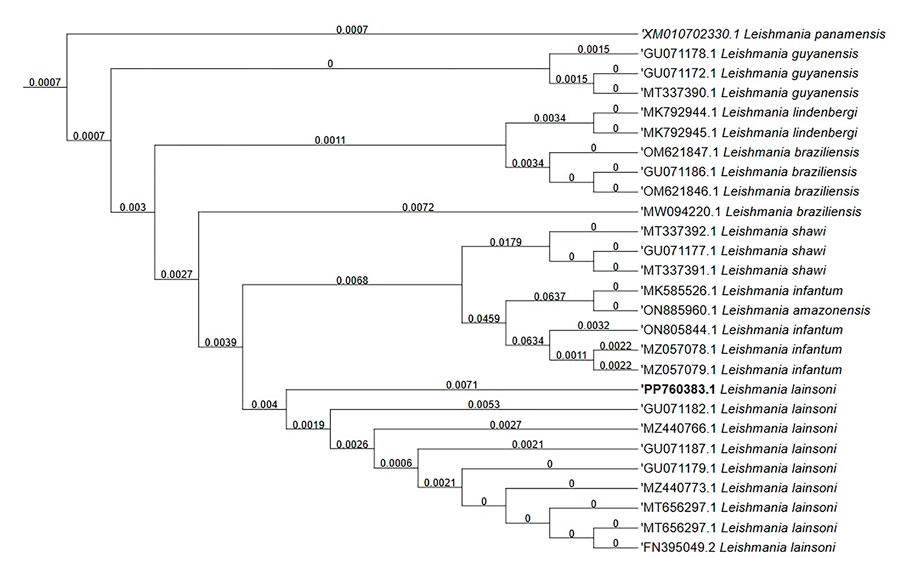
Figure 2. Evolutionary analysis of autochthonous Leishmania (Viannia) lainsoni in dog (Canis familiaris), Rio de Janeiro state, Brazil, 2023. Bold text indicates isolate from this study. Evolutionary history was inferred by using the maximum-likelihood method and Kimura 2-parameter model. The bootstrap consensus tree inferred from 1,000 replicates represents evolutionary history of the taxa analyzed. Branches corresponding to partitions reproduced in <50% bootstrap replicates are collapsed. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) are shown next to the branches. Initial tree(s) for the heuristic search were obtained by applying the BioNJ (https://bionj.org) method to a matrix of pairwise distances estimated by using the maximum composite likelihood approach. This analysis involved 33 nucleotide sequences. There were a total of 463 positions in the final dataset. Evolutionary analyses were conducted in MEGA11 (https://www.megasoftware.net). MEGA used the first position for each codon in the construction of the phylogenetic tree. GenBank accession numbers are shown.
1These authors contributed equally to this article.