Volume 29, Number 8—August 2023
Research
Prospecting for Zoonotic Pathogens by Using Targeted DNA Enrichment
Figure 2
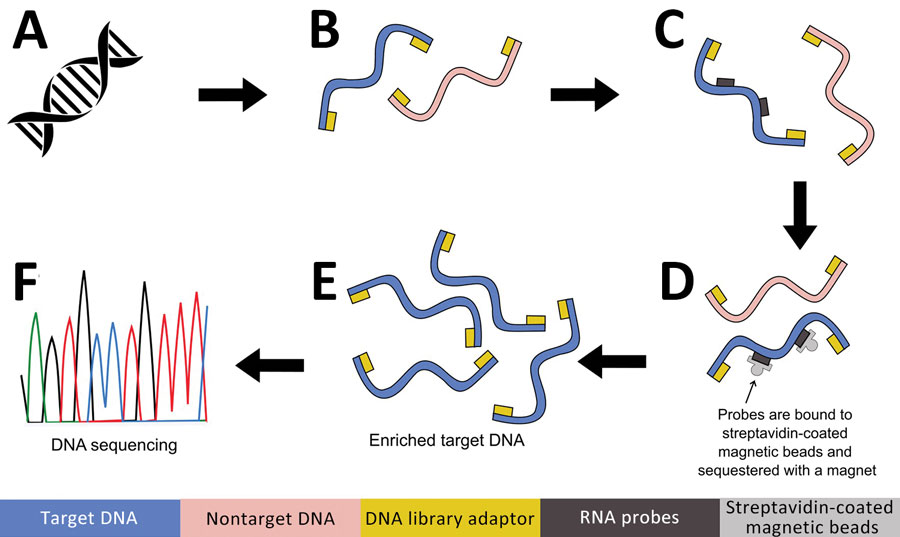
Figure 2. Targeted DNA enrichment workflow for study of prospecting for zoonotic pathogens by using targeted DNA enrichment. A) Genomic DNA extracted using the DNeasy Kit (QIAGEN, https://www.qiagen.com). B) Next-generation sequencing libraries prepared using KAPA Hyperplus Kit (https://www.biocompare.com) and barcoding each library with IDT xGen Stubby Adaptor-UDI Primers (https://www.idtdna.com). C) RNA probes hybridization using the high sensitivity protocol of myBaits version 5. (https://arborbiosci.com). D) Probes bound to streptavidin-coated magnetic beads and sequestered with a magnet (E) 15 cycles PCR amplification of enriched libraries. F) Libraries sequenced on an Illumina Hi-Seq 2500 platform (https://www.illumina.com).
Page created: June 07, 2023
Page updated: July 20, 2023
Page reviewed: July 20, 2023
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.