Volume 30, Number 9—September 2024
Research
Lower Microscopy Sensitivity with Decreasing Malaria Prevalence in the Urban Amazon Region, Brazil, 2018–2021
Figure 4
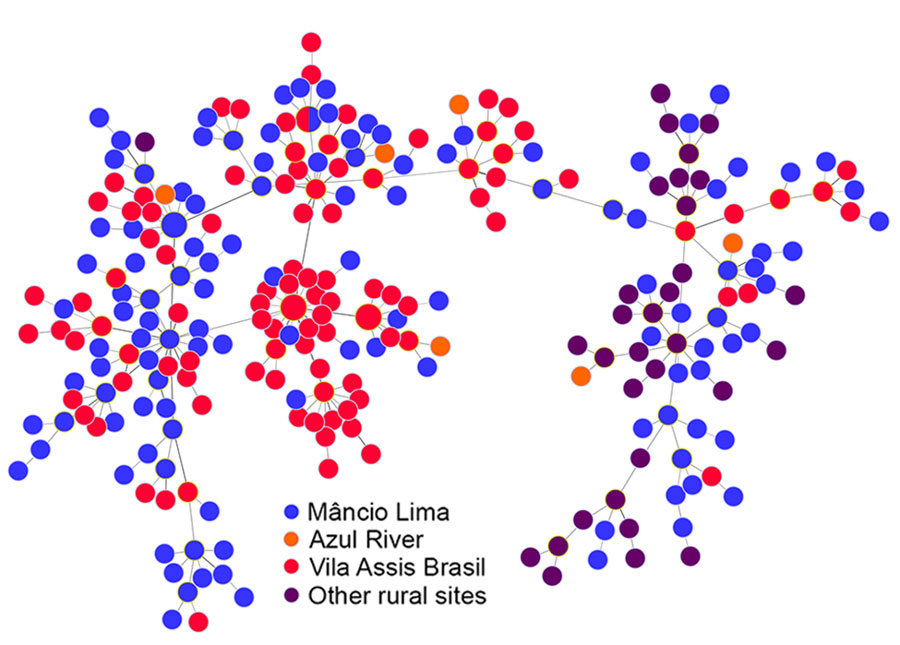
Figure 4. Minimal spanning trees representing the connectivity of Plasmodium vivax haplotypes from the Juruá Valley region of Brazil (264 isolates collected 2016‒2021). Circles represent haplotypes with size linearly proportional to the number of isolates sharing them. Lines connect pairs of haplotypes with <5 allele mismatches and the overall network represents the most likely haplotype genealogy, ensuring that the summed distance of all links of the tree is the minimum possible. Haplotype colors indicate the likely site of infection (Appendix Figure 2). The circle with blue and red slices indicates a haplotype that was shared by 2 parasites from different (urban and rural) origins.
1Group members are listed at the end of this article.
Page created: August 01, 2024
Page updated: August 20, 2024
Page reviewed: August 20, 2024
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.