Disclaimer: Early release articles are not considered as final versions. Any changes will be reflected in the online version in the month the article is officially released.
Volume 31, Number 5—May 2025
Dispatch
High Prevalence of Influenza D Virus Infection in Swine, Northern Ireland
Suggested citation for this article
Abstract
We detected influenza D virus in multiple swine herds in Northern Ireland. Whole-genome sequencing showed several circulating genotypes and novel mutations in the receptor-binding site and esterase domains of the hemagglutinin-esterase fusion protein. Transmission routes of influenza D virus to swine remain to be clarified but could be direct or indirect.
Cattle are the main reservoir of influenza D virus (IDV) worldwide, although the virus is occasionally detected in other species, including swine (1). In Europe, surveillance for IDV in swine has either failed to detect the virus by molecular methods or detected the virus at low prevalence, at <5.6% at the herd level (2). Cattle in Northern Ireland have previously tested positive for IDV (3). Here, we describe detection of IDV in multiple swine herds from Northern Ireland and report on the genetic characterization of the swine-origin IDV strains.
Veterinarians visited 17 swine breeding units, composed of a mixture of farrow-to-wean and farrow-to-finish operations, during January–May 2023. The farms were involved in an ongoing regional control program for porcine reproductive and respiratory syndrome virus (PRRSV) that targeted 10- to 12-week-old pigs according to the PRRSV testing algorithm (https://www.cafre.ac.uk/wp-content/uploads/2023/10/Area-Regional-Control-Final-Report.pdf) for growing pigs. In addition to serum samples for PRRSV testing, 30 nasal swab samples per unit were obtained from the same cohort of growing pigs and tested for the presence of influenza A virus (IAV) and IDV by real-time reverse transcription PCR (rRT-PCR), as previously described (4,5). Serum samples were sent to a commercial diagnostics company (aCare Lab, https://acarelab.com) for determination of PRRSV status by rRT-PCR and open reading frame 5 gene sequencing. Farms were sampled on 1 occasion, except for farm 2170, which was originally sampled in February and resampled in May at the request of the farmer because of a deteriorating clinical situation.
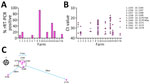
Samples from 7 units (41.2% of herds) tested positive for IDV and had sample positivity of 10%%–93% (Figure 1, panel A). Farm 2170 tested positive for IDV at both timepoints. Associated rRT-PCR cycle thresholds ranged from 17.4 to 36.9 (median 28.0) (Figure 1, panel B). The highest rates of sample positivity were 93% for farm 2163 and 50% for farm 2170; the lowest average cycle thresholds were 28.4 for farm 2163 and 25.1 for farm 2170. Four of 7 IDV-positive farms were also positive for IAV, and 3 of 7 were positive for a field strain of PRRSV (Table). Although clinical signs such as coughing and fever were not recorded at the time of sampling, we retrospectively asked producers whose farms tested positive for IDV about the clinical situation on the farm. Three farms (2160, 2163, 2170) reported increased death and slow growth, 3 (2155, 2156, 2166) reported no overt clinical signs, and no response was received from 1 farm (2179) (Table). A follow-up investigation with the attending veterinarians revealed that 5 of the positive farms (2160, 2163, 2168, 2170, 2179) also kept cattle, whereas 2 farms (2155, 2156) were swine-only farms. Cattle grazed in pastures adjacent to the IDV-positive swine-only units, and >1 worker on each swine unit also worked on the nearby cattle farms. Five of the IDV-positive units were located in a swine-dense region, within a 5.5-km radius (2160, 2163, 2168, 2156, 2170), whereas 2 farms were outside that region: 29 km to the southwest (farm 2155) and 78 km to the southeast (farm 2179) (Figure 1, panel C; Appendix).
We compared hemagglutinin-esterase fusion (HEF) amino acid sequences from swine-origin IDV from Northern Ireland with cattle isolate D/bovine/Northern_Ireland/24280/2017 and noted the substitutions (Figure 2). We used amino acid numbering, as previously described, after removal of the first 16 N-terminal residues (6). We determined the degree of conservation at each substituted position by alignment of all 169 publicly available sequences (Appendix).
We mapped the location of the amino acid substitutions in the HEF protein structure onto a homology model of D/bovine/Northern_Ireland/24280/2017 HEF (Figure 3). Swine-origin strains differed from the bovine sequence at 5 positions around the receptor binding site. Of those positions, 3 were at highly conserved positions: G194R/K, I270M, and K276R. We only detected substitutions at 270 and 276 in samples from farm 2160, whereas we detected G194R/K in samples from 3 farms (2156, 2163, 2170). Swine-origin strains also differed from the bovine sequence at 2 positions around the esterase domain at the highly conserved positions A68V and A324S. We only detected substitutions at 68 in samples from farm 2163, whereas we detected A324S in samples from 3 farms (2160, 2168, 2179).
Interspecies transmission from cattle, followed by some host adaptation and intraspecies spread, most likely initiates swine infection by IDV (7,8). Previous detection of IDV in swine have been associated with sample positivity rates mostly <1% (2). Combined with the low prevalence of IDV at the herd level, that positivity rate suggests intraspecies transmission is limited in swine.
The organization of the agricultural sector in Northern Ireland may partly explain the high prevalence of IDV shown in our study. Multispecies farms containing both cattle and pigs are common and increase the likelihood of multiple IDV spillover events from cattle reservoirs. Indeed, phylogenetic analysis of swine-origin IDV in Northern Ireland identified several distinct genotypes, supporting the idea of repeated introductions from cattle (Appendix). However, the high rate of sample positivity observed on some farms is indicative of efficient pig-to-pig transmission. Furthermore, the continued detection of IDV on 1 farm (2170), 3 months after the initial detection, may represent continuous circulation of the virus within the herd. Those findings provide preliminary evidence for efficient adaptation of IDV to swine hosts and establishment within those populations. In addition, our data suggest that spread between farms might be a factor contributing to the high herd-level prevalence observed. The close genetic relatedness of some IDV strains isolated from both neighboring and geographically distant farms may indicate both local (direct) and long-distance (indirect) spread. Although common sources of indirect spread such as contaminated service vehicles should be considered, we must also consider cattle movements as a possible source of spread between swine units.
The molecular basis of IDV host adaptation has yet to be determined but likely includes changes that affect receptor binding. The receptor binding site in the IDV HEF protein is in a shallow cavity surrounded by secondary structure elements from the 170, 190, and 270 loops and the 230 helix (6). Substitutions in those secondary structure elements have the potential to modify receptor binding activity. For example, the open channel between the 230 helix and 270 loop is thought to enable broad cell tropism of IDV. Substitutions in those elements in the related influenza C virus HEF close that channel and restrict broad cell tropism (6). Candidate swine adaptations A236V and R268K have been previously identified after likely cattle-to-pig transmission (7), and L100F was observed after experimental infection of swine with a bovine-origin IDV isolate (9). In our study, we identified several additional candidates in both the receptor binding site and esterase. Of particular interest is mutation of G194 to a basic residue (R or K). Sequences from farms with the highest sample positivity rates (2163 and 2170) contained that mutation, and phylogenetic analysis indicated that strains from those farms were only distantly related and therefore seem to have independently converged on that adaptation. However, multiple routes to swine adaptation for IDV are likely, and alternative substitutions, such as the 270–273–276 triplet and the esterase changes at positions 68 and 324, should be examined further. Researchers should validate the proposed swine adaptive mutations, including receptor binding analysis and experimental transmission studies. Transmission routes of IDV to swine remain to be clarified but could be direct (e.g., contact with infected cattle) or indirect (e.g., contaminated vehicles).
Dr. Lemon is a researcher in the Mammalian Virology Unit at the Agri-Food and Bioscience Institute in Northern Ireland. His research interests include the molecular epidemiology of animal influenza viruses.
Acknowledgment
We thank the swine producers and veterinarians from Northern Ireland for their support.
References
- Kwasnik M, Rola J, Rozek W. Influenza D in domestic and wild animals. Viruses. 2023;15:2433. DOIPubMedGoogle Scholar
- Gaudino M, Chiapponi C, Moreno A, Zohari S, O’Donovan T, Quinless E, et al. Evolutionary and temporal dynamics of emerging influenza D virus in Europe (2009-22). Virus Evol. 2022;8:
veac081 . DOIPubMedGoogle Scholar - Dane H, Duffy C, Guelbenzu M, Hause B, Fee S, Forster F, et al. Detection of influenza D virus in bovine respiratory disease samples, UK. Transbound Emerg Dis. 2019;66:2184–7. DOIPubMedGoogle Scholar
- Hause BM, Ducatez M, Collin EA, Ran Z, Liu R, Sheng Z, et al. Isolation of a novel swine influenza virus from Oklahoma in 2011 which is distantly related to human influenza C viruses. PLoS Pathog. 2013;9:
e1003176 . DOIPubMedGoogle Scholar - Lagan P, Hamil M, Cull S, Hanrahan A, Wregor RM, Lemon K. Swine influenza A virus infection dynamics and evolution in intensive pig production systems. Virus Evol. 2024;10:
veae017 . DOIPubMedGoogle Scholar - Song H, Qi J, Khedri Z, Diaz S, Yu H, Chen X, et al. An open receptor-binding cavity of hemagglutinin-esterase-fusion glycoprotein from newly identified influenza D virus: basis for its broad cell tropism. PLoS Pathog. 2016;12:
e1005411 . DOIPubMedGoogle Scholar - Gorin S, Richard G, Hervé S, Eveno E, Blanchard Y, Jardin A, et al. Characterization of influenza D virus reassortant strain in swine from mixed pig and beef farm, France. Emerg Infect Dis. 2024;30:1672–6. DOIPubMedGoogle Scholar
- Kaplan BS, Falkenberg S, Dassanayake R, Neill J, Velayudhan B, Li F, et al. Virus strain influenced the interspecies transmission of influenza D virus between calves and pigs. Transbound Emerg Dis. 2021;68:3396–404. DOIPubMedGoogle Scholar
- Gorin S, Richard G, Quéguiner S, Chastagner A, Barbier N, Deblanc C, et al. Pathogenesis, transmission, and within-host evolution of bovine-origin influenza D virus in pigs. Transbound Emerg Dis. 2024;2024:
9009051 . DOIGoogle Scholar
Figures
Table
Suggested citation for this article: Lagan P, Lemon K. High prevalence of influenza D virus infection in swine, Northern Ireland. Emerg Infect Dis. 2025 May [date cited]. https://doi.org/10.3201/eid3105.241948
Original Publication Date: March 26, 2025
Table of Contents – Volume 31, Number 5—May 2025
| EID Search Options |
|---|
|
|
|
|
|
|


Please use the form below to submit correspondence to the authors or contact them at the following address:
Ken Lemon, Veterinary Sciences Division, Agri-Food and Biosciences Institute, Stormont Stoney Road, Belfast BT4 3SD, Northern Ireland
Top