Disclaimer: Early release articles are not considered as final versions. Any changes will be reflected in the online version in the month the article is officially released.
Volume 31, Number 4—April 2025
Research
Carbapenem-Resistant, Virulence Plasmid–Harboring Klebsiella pneumoniae, United States
Suggested citation for this article
Abstract
Carbapenem-resistant and virulence plasmid–harboring Klebsiella pneumoniae (pVir-CRKP) has emerged and spread globally, yet clinical investigations from the United States remain limited. We conducted a genomic analysis of 884 unique carbapenem-resistant K. pneumoniae isolates from a multicenter US cohort and identified 6 pVir-CRKP isolates, including 2 sequence type (ST) 23, 2 ST893, and 2 ST11 isolates. Patients infected with pVir-CRKP experienced high Pitt bacteremia scores and a 33% 30-day mortality rate. The pVir-CRKP isolates exhibited significant sequence variation in virulence genes and plasmids, along with differences in mucoviscosity, capsule production, survival in normal human serum, resistance to killing by human polymorphonuclear neutrophils, and in vivo pathogenicity. Phylogenetic analyses showed that most pVir-CRKP isolates were genetically similar to strains reported from other global regions. The emergence of pVir-CRKP with higher virulence potential and carbapenem resistance in the United States than the predominant carbapenem-resistant K. pneumoniae clone underscores the need for active global surveillance.
Carbapenem-resistant Klebsiella pneumoniae (CRKP) is a major concern to global public health because it is highly resistant to most β-lactam and β-lactamase inhibitors (1). Strains termed hypervirulent K. pneumoniae (HvKP) cause community-acquired invasive infections in healthy persons (2). The emergence of convergent strains of K. pneumoniae, possessing features of both HvKP and CRKP, has elevated the concern by linking virulence and resistance in a single strain (3). The World Health Organization warned of a global increase in both HvKP and CRKP (4).
The convergent strains are reported mostly from East Asia but also from other global regions. Several previous studies documented the acquisition of carbapenemase-encoding plasmids in HvKP strains. Karlson et al. (5) reported a blaKPC-2–carrying sequence type (ST) 23 HvKP isolate from the United States, and Chen et al. (6) reported 18 blaKPC-2–carrying ST23, ST65, and ST86 HvKP isolates from Singapore. In addition, Beyrouthy et al. (7) reported a blaOXA-48–harboring ST86 HvKP isolate from France. The movement of virulence plasmids into carbapenem-resistant strains has also been reported. In 2018, Gu et al. (8) identified 5 virulence plasmid–harboring ST11 CRKP strains from China with enhanced virulence (compared with strains lacking the plasmid); more recently, Zhou et al. (9) found that a large proportion of ST11 CRKP strains from China have acquired virulence genes or plasmids.
The virulence of HvKP isolates is attributed mainly to a pK2044-like plasmid (pVir) that harbors genes encoding mucoid regulators and siderophores (10). Mucoid regulators, including RmpADC and RmpA2, enhance capsule production and mucoviscosity of HvKP strains, increasing resistance to neutrophil phagocytosis and serum complement-mediated killing (11). Siderophores, which are small molecules secreted by bacteria to accumulate iron, include pVir-encoded aerobactin (iuc) and salmochelin (iro) (12), as well as 2 chromosome-encoded siderophores, yersiniabactin (ybt) and colibactin (clb), which also contribute to virulence (12). Nevertheless, genotypic prediction of HvKP virulence remains a challenge because the presence of virulence genes does not always correlate with increased virulence in vivo. Russo et al. indicated that the combination of 5 pVir-borne virulence factors, namely iucA (from the iuc gene cluster), iroB (from the iro gene cluster), peg-344 (a metabolite transporter), rmpA (from the rmpADC gene cluster), and rmpA2, had an overall accuracy of 94% in predicting the hypervirulence phenotype of HvKP in an animal infection model (13).
In this study, we analyzed the genomic features of carbapenem-resistant and virulence plasmid–harboring K. pneumoniae (pVir-CRKP) isolates from US cohorts associated with the international multicenter prospective Consortium on Resistance Against Carbapenem in Klebsiella pneumoniae and other Enterobacterales 2 (CRACKLE-2) study (14,15). We describe the clinical characteristics of pVir-CRKP isolates and conducted a comprehensive phenotypic analysis to assess their in vitro, ex vivo, and in vivo virulence, and tracked the phylogenetic origins of those pVir-CRKP strains.
Study Cohort
CRACKLE-2 (ClinicalTrials.gov no. NCT03646227) consecutively enrolled hospitalized patients with carbapenem-resistant Enterobacterales (CRE) isolates from all anatomic sources during 2016–2018 (14,15). In our analysis, patients from the United States were eligible for inclusion when their first qualifying CRE culture was positive for K. pneumoniae. The Institutional Review Board of each study site approved the study. We evaluated Charlson comorbidity index (CCI) (16), Pitt bacteremia score (17), and desirability of outcome ranking (DOOR) (14) as previously described.
Quantifying Mucoviscosity and Capsule Production
We measured mucoviscosity as previously described (18). In brief, we adjusted overnight cultures to a 600-nm optical density (OD600) of 1 and then centrifuged. We measured the OD600 values (hypermucoid index) in 3 samples. We quantified the capsule using the uronic acid method, as previously described (19) (Appendix)
Whole-Genome Sequencing and Bioinformatics Analysis
We performed whole-genome sequencing (WGS) as previously described using an Illumina NextSeq 500 platform (https://www.illumina.com) with 2 × 150 bp paired-end reads (15). We assembled draft genomes using SPAdes version 3.13.0 as described (20). We used Kleborate version 2.3 (21) to analyze sequence type, capsular type, rmpADC, rmpA2, ybt, clb, iuc, and iro. We detected peg-344 by BLASTn version 2.16.0 (ftp://ftp.ncbi.nlm.nih.gov/blast/executables/blast+) against the gene sequence (GenBank accession no. BAH65947.1). We identified resistance genes by AMRFinderPlus version 3.10.20 (22), and ARIBA version 2.14.6 (23). We considered isolates with contig sequences showing >95% BLASTn identity and >65% sequence coverage to pK2044 (accession no. AP006726.1), and harboring iuc/iro and rmpADC/A2 as pVir-KP. We sequenced those pVir-KP isolates with Oxford Nanopore (https://nanoporetech.com) and hybrid assembled them with short reads to closure via Unicycler version 0.4.9 (24). We visualized linear alignment using Easyfig version 2.1 (25) and used Prism 10 for Windows 64-bit version 10.2.3 (GraphPad Software, www.graphpad.com) for data analysis.
Serum Bactericidal Activity and Polymorphonuclear Neutrophil Phagocytosis Assays
We determined serum bactericidal activity as described (26). In brief, we combined 5 × 105 colony-forming units (CFU) of bacteria with normal human serum (NHS) and RPMI/H medium in an assay. We rotated assay tubes, diluted serially, and plated on LB agar plates. We calculated percentage survival as CFUwith NHS/CFUwithout NHS × 100. We isolated human polymorphonuclear neutrophils (PMNs) using a standard method (27) and determined PMN killing as previously described (26). We combined, incubated, and diluted bacteria (2.5 × 106 CFU), PMNs (5 × 105 CFU in RPMI/H), and NHS, then plated them on LB agar and enumerated colonies the next day (Appendix). We obtained blood from healthy participants in accordance with a protocol approved by the Institutional Review Board for human subjects at the US National Institutes of Health (protocol no. 01IN055).
Animal Infection Models
We used a well-established murine intranasal infection model for this study (28). We anesthetized female BALB/c mice by intraperitoneal injection. We placed a total of 50 μL of K. pneumoniae suspension (≈104 CFUs) on the nares of mice (25 μL per nare) for aspiration into the lungs. We quantified bacterial burdens of each tissue by serial dilutions on LB agar plates (Appendix). We used K. pneumoniae strains ATCC43816 (ST493/capsular locus [KL] 2) and NJST258_2 (ST258/KL107) as controls for comparison. Hackensack Meridian Health Institutional Animal Care and Use Committee approved all vertebrate animal experiments.
Carbapenem-Resistant K. pneumoniae Isolates
We illustrated DOOR outcomes (Figure 1, panel A), along with the virulome, resistome, and porin mutations associated with different STs (Figure 1, panel B), for 884 CRKP isolates. Among them, 385 (43.6%) isolates possessed antimicrobial resistance genes and >1 of ybt (n = 384), clb (n = 224), iuc (n = 25), iro (n = 13), rmpA (n = 4), rmpA2 (n = 5), or peg-344 (n = 4) genes. CRKP isolates harboring 2 virulence genes (ybt and clb) and genes encoding resistance to aminoglycosides, fluoroquinolones, sulfonamides, trimethoprim, sulfamethoxazole, β-lactam and carbapenem were the most common (n = 56) (Figure 1, panel C). ST258 isolates with chromosomal ybt, clb, or both were the most common (237/385 [61.6%]) among virulence gene–harboring CRKP isolates (Figure 1, panel D). For 25 CRKP isolates with >1 of the 5 pVir-borne virulence genes used to predict HvKP in the Russo et al. study (13), ST258 (n = 10) and ST231 (n = 8) with iuc/iro were the most common. However, only 6 isolates displaying >95% BLASTn identity and >65% sequence coverage compared with pK2044 were pVir-CRKP strains; 2 were ST23, 2 ST11, and 2 ST893 isolates (Figure 1, panel E). ST23 and ST11 isolates harbored all 5 virulence markers (13); however, many of those operons and genes were not intact. The remaining 19 CRKP isolates contained plasmidborne iuc but showed low coverage (<8%) compared with pK2044 and were negative for rmpA/A2. Of those 19 isolates, 8 (>75%) displayed high coverage compared with the iuc-harboring IncFII(pAMA1167-NDM-5) plasmid pC346_2 (GenBank accession no. CP067712.1) from K. pneumoniae, whereas the others exhibited high coverage (> 75%) compared with the iuc-harboring IncFIB-IncFIC (FII) plasmid p3PCN033 (GenBank accession no. CP006635.1) from E. coli (29). In addition, all 10 ST258 strains harbored the iro operon in an ICEKp10 element on the chromosome.
Clinical Characterization of pVir-CRKP–Infected Patients
We cultured the 6 US pVir-CRKP isolates from respiratory (n = 3), urine (n = 2), or wound (n = 1) specimens from patients in Cleveland (n = 1) and Houston (n = 1) in 2016 and in Atlanta (n = 4) during 2016 and 2017. Four pVir-CRKP isolates were hospital acquired, and 2 were community acquired. Apart from K. pneumoniae, no other pathogens were cultured from the clinical specimens. Five of the patients were middle-aged (median age 56 [IQR 55–60] years), and 5 were male. Of note, patients infected with pVir-CRKP had lower CCI scores (median 2 [IQR 1–4] vs. median 3 [IQR 1–5]) but higher Pitt bacteremia scores (median 3.5 [IQR 2–6] vs. median 3 [IQR 1–5]) compared with patients infected with CRKP (15), which suggests that pVir-CRKP isolates possess increased virulence. Patient cases associated with pVir-CRKP demonstrated a 30-day mortality rate of 33%. In contrast, in all the US patients with CRKP infections, unadjusted 30-day all-cause mortality rate was 23% (15). However, the small sample size of pVir-CRKP–infected patients may limit the reliability of the observed mortality rate. Two patients with CCI scores >3 died, whereas patients with CCI scores <3 survived without adverse events (Table 1).
Genomic and Microbiological Characterization of pVir-CRKP
Genomic analysis classified ARLG-4622 and ARLG-3254 as ST23 with capsule type KL1 and lipopolysaccharides type O1, ARLG-4744 and ARLG-7683 as ST11 with KL64 and O1, and ARLG-3484 and ARGL-4720 as ST893 with KL20 and O3. Capsule polysaccharide (CPS) synthesis genes UDP-glucose dehydrogenase (ugd) and acid phosphatase (cpsACP) were truncated by frameshift mutations, ugd in ARLG-4622 and cpsACP in ARLG-3254 (Table 2).
The ST23 pVir-CRKP isolates harbored blaKPC-2 on a repA_KPC-2 plasmid (ARLG-4622) or on a pQIL-like IncFIIK2 plasmid (ARLG-3254). The ST893 isolates encoded extended-spectrum β-lactamase (ESBL) CTX-M-15 and carbapenemase OXA-48 on a complex recombinant plasmid with IncL/M, IncFII and IncFIA(HI1) replicons. Of note, truncations in the chromosomal colistin resistance associated gene mgrB, and insertion sequence (IS) interruptions of porins (ompK35 and ompK36) were detected in both ST893 isolates. Although the ST11 isolates were carbapenem resistant (Table 3), no carbapenemase was identified. Further analysis revealed that the ST11 isolates contain the ESBL gene blaCTX-M-65, and nonfunctional OmpK35 (IS interruption) and OmpK36 (glycine-aspartate insertion) (Table 2). In addition, we detected other antimicrobial resistance genes, including fosA3, qnrS1, sul2, and dfrA14, on ESBL or carbapenemase plasmids.
The pVir-CRKP isolates displayed multidrug resistance; their resistance genotype correlated with phenotypic resistance. For example, the ST23 and ST893 isolates encoding carbapenemase were resistant to carbapenems and cephalosporins, whereas ST11 isolates with ESBL and porin disruptions were resistant to ertapenem and cephalosporins but susceptible to imipenem and meropenem. The pVir-CRKP isolates with QRDR (gyrA and parC) mutations and quinolone resistance genes (qnrS1 and qnrB1) exhibited resistance to quinolones. The ST11 pVir-CRKP isolates contained fosA3 and were resistant to fosfomycin; the ST893 isolates with MgrB disruptions were resistant to colistin (Table 3). All isolates exhibited susceptibility to ceftazidime/avibactam and cefiderocol.
The size of pVir in the 6 pVir-CRKP isolates was 185.4–229.8 Kb (Figure 2). The pVir in ST23 isolates (ARLG-4622 and ARLG-3254) had high sequence similarity (>99% BLAST identity and >97% coverage) to the hypervirulence plasmid pK2044 (GenBank accession no. AP006726.1) and contained intact operons encoding iro (iroBCDN) and iuc (iutA-iucABCD) and a truncated rmpA2. The rmpA operon (rmpADC) was intact in ARLG-3254; however, most of the operon was truncated in ARLG-4622 (except for rmpD). pVir in ST11 isolates (ARLG-4744 and ARLG-7683) also displayed high sequence similarity (>99% BLAST identity and ≈92% coverage) to pK2044 and contained iuc and rmpDC but had truncated iro and rmpA2. The rmpA locus was intact in ARLG-4744, but was interrupted by an IS in ARLG-7683. In contrast, the pVir from ST893 isolates (ARLG-3484 and ARLG-4720) showed lower similarity (>99% BLAST identity and ≈67% coverage) to pK2044 with intact iuc and truncated rmpA2; the iro and rmpADC loci were lost. All virulence plasmids possessed IncFIB(K) and IncHI1B replicons.
Mucoviscosity and Capsule Production
We next assessed mucoviscosity and CPS production with the 6 pVir-CRKP isolates and correlated the results with the presence of the rmpADC operon. The hypermucoid index for all the pVir-CRKP isolates was significantly lower than that for the reference strain ATCC43816. Isolates with intact rmpADC (ARLG-3254 and ARLG-4744) exhibited significantly higher mucoid indices than isolates with a truncated rmpADC (ARLG-4622 and ARLG-7683) or those missing rmpADC (ARLG-3484 and ARLG-4720). For example, the hypermucoid index for ARLG-3254 (0.53 + 0.03) was significantly greater than that for ARLG-4622 (0.31 + 0.01) (p<0.01) (Figure 3, panel A).
CPS production and mucoviscosity are 2 different properties of K. pneumoniae with overlapping and confounding phenotypes (30). In our study, CPS production was independent of the presence of intact rmpADC or rmpA2, and correlated with each isolate’s genetic background and capsule type, because ST23-KL1 isolates produced the greatest amount of CPS (19.84 + 1.65 µg/OD600), followed by ST893-KL20 isolates (16.40 + 2.74 µg/OD600), and ST11-KL64 isolates (13.36 + 2.96 µg/OD600) (Figure 3, panel B).
Differential Survival of pVir-CRKP Isolates
To assess the ability of the US pVir-CRKP isolates to circumvent killing by the human innate immune system, we tested survival of the 6 selected clinical isolates in NHS. Our previous study demonstrated that clinical K. pneumoniae strains without pVir usually have <40% survival in 25% NHS and <20% survival in 83% NHS after 1 hour (26). We found that the pVir-CRKP isolates (except for ARLG-4622) had >75% (96.1% + 16.0%) survival in 25% NHS and >50% (76.9% + 24.9%) survival in 83% NHS after 1 hour (Figure 4, panels A, B), indicating those isolates have enhanced capacity to survive in NHS by comparison.
Although the pVir-CRKP isolates (except ARLG-4622) displayed greater survival in NHS, they exhibited varied survival during phagocytic interaction with human PMNs (Figure 4, panels C, D). For example, survival of ARLG-3254 (ST23), ARLG-3484 (ST893) and ARLG-4720 (ST893) was not notably lower after 60 min of phagocytic interaction with PMNs. By comparison, survival of ARLG-4744 (ST11) and ARLG-7683 (ST11) decreased significantly by interaction with PMNs (29% + 14% vs. 26% + 15%; p<0.05).
Virulence of pVir-CRKP Isolates in Mouse Pneumonia Model
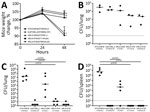
We evaluated the in vivo virulence of pVir-CRKP using a mouse pneumonia model. Weight of the mice (measured by weight relative to the 0 h baseline) inoculated with HvKP strains, ATCC43816 (virulent control strain) and the ARLG-3254 (ST23) clinical isolate, was significantly reduced (p<0.01) compared with weight of mice inoculated with a clinical K. pneumoniae ST258 control strain (NJST258_2) or the other isolates tested (Figure 5, panel A). At 2 hours postinfection, we observed no significant difference in viable bacteria recovered from the lungs and spleens of mice inoculated with 3 pVir-CRKP isolates and 2 control strains (Figure 5, panel B). However, at 48 hours postinfection, the bacterial load in the lungs (Figure 5, panel C) and spleens (Figure 5, panel D) in mice infected with ATCC43816 and ARLG-3254 was significantly higher (p<0.01) than that in mice infected with NJST258_2, ARLG-4744 and ARLG-4720, suggesting ARLG-3254 (ST23) has significantly higher virulence in vivo than NJST258_2 and 2 other pVir-CRKP isolates at this inoculum.
Phylogenetic Analyses of pVir-CRKP Isolates
To track the possible origin of US pVir-CRKP isolates, we performed phylogenetic analyses with additional genomes of the same ST from the National Center for Biotechnology Information Genome database (https://www.ncbi.nlm.nih.gov/datasets/genome). We aligned the top 50 genetically related genomes and the ARLG-4622 genome sequence to the ST23 reference genome ARLG-3254 (GenBank accession no. CP067777), identifying 5,037 core-genome SNPs (cgSNPs). The ARLG-3254 and ARLG-4622 differed by 280 pairwise cgSNPs (Appendix Figure 1). ARLG-3254 (from Houston, Texas, USA) was most closely related to the ST23 strains isolated in the United States, whereas ARLG-4622 (from Cleveland, Ohio, USA) was more similar to strains reported in China (Appendix Figure 1). Both ST23 isolates showed long phylogenetic distance from the first KL1-ST23 pVir-CRKP strain reported in the United States (5) (Appendix Figure 1). For the ST11 strains, the top 50 genetically related genomes and the ARLG-7683 genome sequence were aligned to the reference genome ARLG-4744 (GenBank accession no. CP139237); we identified 1,023 cgSNPs. The ARLG-4744 and ARLG-7683 collected in Atlanta were genetically similar (differing by 11 pairwise cgSNPs) and most closely related to strains collected in China (Appendix Figure 2). For the ST893 strains, all 18 publicly available genomes and the ARLG-3484 genome sequence were aligned to the ST893 reference genome ARLG-4720 (GenBank accession no. CP139225); we detected 1,466 cgSNPs. The ARLG-4720 and ARLG-3484 from Atlanta differed by only 6 pairwise cgSNPs and were most similar to strains collected in Iran in 2018 (Appendix Figure 3).
In this study, we analyzed the genomic features of 884 CRKP isolates from CRACKLE-2 study US cohorts and identified 6 (0.7%) pVir-CRKP isolates. The 6 pVir-CRKP isolates belonged to 3 different STs, and each isolate exhibited enhanced production of CPS, survival in NHS and resistance to PMN killing compared with the well-characterized and virulence plasmid–lacking classical K. pneumoniae strain NJST258_2 (the predominant CRKP clone in the United States). Phylogenetic analyses revealed that 5 of 6 US pVir-CRKP isolates were genetically similar to strains reported from China and Iran.
pVir-CRKP strains have mostly been reported from East Asia. Here, we report the emergence of pVir-CRKP strains in the United States during 2016–2017. The first reported US pVir-CRKP strain (DHQP1701672) was collected in 2016 from a 65-year-old patient after travel to South America (5). DHQP1701672 is an ST23 strain harboring pVir and pKpQIL-like blaKPC-2 plasmid. Kochan et al. (31) screened the genomes of 2,608 multidrug-resistant K. pneumoniae isolates from 3 different US studies and identified 47 isolates containing >1 HvKP virulence gene. In this study, we identified 2 ST23 pVir-CRKP isolates; 1 harbored a pKpQIL-like blaKPC-2 plasmid. However, we noted significant phylogenetic distance between those isolates and DHQP1701672, indicating that they did not evolve from DHQP1701672. One ST23 pVir-CRKP isolate showed close genetic distance to an HvKP strain reported in the United States. Given that this pVir-CRKP isolate harbored a pKpQIL-like blaKPC-2 plasmid, which is prevalent in North America (32), we hypothesize that the isolate may have arisen from the acquisition of a pKpQIL-like blaKPC-2 plasmid by a local US ST23 HvKP. Another ST23 pVir-CRKP isolate showed close phylogenetic distance to the strains reported in China. In addition, we identified ST11 and ST893 pVir-CRKP isolates. ST11 is the most common clone of CRKP and pVir-CRKP in China (9), and the 2 ST11 pVir-CRKP isolates identified here were phylogenetically similar to strains from China. Last, ST893 is a minor clone mainly reported in China (33) and Iran (34), and the ST893 pVir-CRKP isolates reported in our study were similar to strains reported from Iran. Overall, our study indicates that, although most US pVir-CRKP isolates were genetically related to strains from other countries, they may also arise domestically through the acquisition of a prevalent circulating carbapenemase gene.
Ex vivo experiments showed that most of the pVir-CRKP isolates displayed increased antiserum capacity in NHS and high antiphagocytosis capacity toward PMNs. Although the ST23 pVir-CRKP isolate ARLG-4622 harbored the 5 virulence genes on pVir (13), it exhibited remarkably reduced antiserum and antiphagocytosis capacities. Further genomic analysis revealed that rmpAC and ugd genes were truncated in ARLG-4622. RmpA amplifies capsule production and mucoviscosity in HvKP strains, thereby enhancing their resistance to neutrophil phagocytosis and killing mediated by serum complement (11). Ugd plays a crucial role in the formation of an antiphagocytic capsule, which serves to protect bacteria from the host’s immune system (35). The defects of RmpA and Ugd potentially reduced the virulence of ARLG-4622, making it more vulnerable to NHS and PMNs. Therefore, it is crucial to assess loss-of-function mutations in virulence genes and CPS synthesis genes when attempting to genetically predict the virulence of K. pneumoniae.
Because our understanding of K. pneumoniae virulence, especially that of multidrug-resistant HvKP strains, is limited, predicting virulence remains a challenge and warrants further investigation. In the mouse pneumonia model, only the ST23 pVir-CRKP isolate demonstrated enhanced virulence; that finding is consistent with the results from a study that found most K. pneumoniae isolates with both multidrug-resistance and virulence genes had low virulence (31). Although ST11 and ST893 pVir-CRKP isolates did not demonstrate enhanced virulence in the mouse in vivo, the enhanced ability of those isolates to avoid killing by human serum and PMNs may confer enhanced survival during human interaction. Such a phenotype could pose a substantial risk to immunocompromised populations, potentially leading to fatal outcomes (8). For example, in our study, the patient infected with ARLG-4744 (ST11) had a high Pitt score and died within 30 days.
A limitation of our study is that the incidence of pVir-CRKP in the United States could be underestimated because we collected only CRKP isolates from hospitalized patients and did not include intestinal colonization samples; nonetheless, a strength of our study is that we used a standardized, contemporary approach to include hospitalized patients with CRKP. Second, although we identified potential clonal transmission within ST11 and ST893 pVir-CRKP isolates, we do not have enough information, especially information regarding environmental samples, to confirm the transmission events. Third, the small sample size of pVir-CRKP–infected patients may limit the accuracy of the observed mortality rate. Finally, the data were somewhat outdated, and a more recent dataset could be more clinically relevant.
In summary, pVir-CRKP with enhanced virulence and carbapenem resistance has emerged in the United States. pVir-CRKP strains exhibited heterogeneous in vitro, ex vivo, and in vivo virulence, warranting further investigation to genetically predict their pathogenicity. Isolates identified in this study were predominantly genetically related to strains from other countries, which highlights the critical importance of global molecular surveillance.
Dr. Jiang is a postdoctoral researcher in infectious diseases and clinical microbiology at the Center for Discovery and Innovation, Hackensack Meridian Health. His main interests focus on the epidemiology of infectious diseases, bacterial pathogenesis, and antimicrobial resistance.
Acknowledgments
This study was supported in part by the National Institute of Allergy and Infectious Diseases, National Institutes of Health (NIAID grant no. R01AI090155), and the Intramural Research Program. Bacterial isolates identified as ARLG were provided by the Antibacterial Resistance Leadership Group Laboratory Center, which is supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (award no. UM1AI104681).
Individual deidentified participant data (and supporting documentation, data dictionaries, and protocol) that underlie the results in this article are available to investigators after submission of a plan for data use, approval by the ARLG or designated entity, and execution of required institutional agreements. Provision might be contingent upon the availability of funding for data preparation and deidentification. More information is at https://arlg.org/how-to-apply/request-data. Sequences are publicly available through the National Center for Biotechnology Information (accession no. PRJNA658369).
References
- Wyres KL, Lam MMC, Holt KE. Population genomics of Klebsiella pneumoniae. Nat Rev Microbiol. 2020;18:344–59. DOIPubMedGoogle Scholar
- Siu LK, Yeh KM, Lin JC, Fung CP, Chang FY. Klebsiella pneumoniae liver abscess: a new invasive syndrome. Lancet Infect Dis. 2012;12:881–7. DOIPubMedGoogle Scholar
- Chen L, Kreiswirth BN. Convergence of carbapenem-resistance and hypervirulence in Klebsiella pneumoniae. Lancet Infect Dis. 2018;18:2–3. DOIPubMedGoogle Scholar
- World Health Organization. Antimicrobial resistance, hypervirulent Klebsiella pneumoniae—global situation. 2024 [cited 2024 Sep 20] https://www.who.int/emergencies/disease-outbreak-news/item/2024-DON527
- Karlsson M, Stanton RA, Ansari U, McAllister G, Chan MY, Sula E, et al. Identification of a carbapenemase-producing hypervirulent Klebsiella pneumoniae isolate in the United States. Antimicrob Agents Chemother. 2019;63:e00519–19. DOIPubMedGoogle Scholar
- Chen Y, Marimuthu K, Teo J, Venkatachalam I, Cherng BPZ, De Wang L, et al. Acquisition of plasmid with carbapenem-resistance gene blakpc2 in hypervirulent Klebsiella pneumoniae, Singapore. Emerg Infect Dis. 2020;26:549–59. DOIPubMedGoogle Scholar
- Beyrouthy R, Dalmasso G, Birer A, Robin F, Bonnet R. Carbapenem resistance conferred by OXA-48 in K2-ST86 hypervirulent Klebsiella pneumoniae, France. Emerg Infect Dis. 2020;26:1529–33. DOIPubMedGoogle Scholar
- Gu D, Dong N, Zheng Z, Lin D, Huang M, Wang L, et al. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis. 2018;18:37–46. DOIPubMedGoogle Scholar
- Zhou K, Xiao T, David S, Wang Q, Zhou Y, Guo L, et al. Novel subclone of carbapenem-resistant Klebsiella pneumoniae sequence type 11 with enhanced virulence and transmissibility, China. Emerg Infect Dis. 2020;26:289–97. DOIPubMedGoogle Scholar
- Russo TA, Carlino-MacDonald U, Drayer ZJ, Davies CJ, Alvarado CL, Hutson A, et al. Deciphering the relative importance of genetic elements in hypervirulent Klebsiella pneumoniae to guide countermeasure development. EBioMedicine. 2024;107:
105302 . DOIPubMedGoogle Scholar - Lai YC, Peng HL, Chang HY. RmpA2, an activator of capsule biosynthesis in Klebsiella pneumoniae CG43, regulates K2 cps gene expression at the transcriptional level. J Bacteriol. 2003;185:788–800. DOIPubMedGoogle Scholar
- Lam MMC, Wick RR, Wyres KL, Gorrie CL, Judd LM, Jenney AWJ, et al. Genetic diversity, mobilisation and spread of the yersiniabactin-encoding mobile element ICEKp in Klebsiella pneumoniae populations. Microb Genom. 2018;4:
e000196 . DOIPubMedGoogle Scholar - Russo TA, Alvarado CL, Davies CJ, Drayer ZJ, Carlino-MacDonald U, Hutson A, et al. Differentiation of hypervirulent and classical Klebsiella pneumoniae with acquired drug resistance. MBio. 2024;15:
e0286723 . DOIPubMedGoogle Scholar - van Duin D, Arias CA, Komarow L, Chen L, Hanson BM, Weston G, et al.; Multi-Drug Resistant Organism Network Investigators. Molecular and clinical epidemiology of carbapenem-resistant Enterobacterales in the USA (CRACKLE-2): a prospective cohort study. Lancet Infect Dis. 2020;20:731–41. DOIPubMedGoogle Scholar
- Wang M, Earley M, Chen L, Hanson BM, Yu Y, Liu Z, et al.; Multi-Drug Resistant Organism Network Investigators. Clinical outcomes and bacterial characteristics of carbapenem-resistant Klebsiella pneumoniae complex among patients from different global regions (CRACKLE-2): a prospective, multicentre, cohort study. Lancet Infect Dis. 2022;22:401–12. DOIPubMedGoogle Scholar
- Charlson M, Szatrowski TP, Peterson J, Gold J. Validation of a combined comorbidity index. J Clin Epidemiol. 1994;47:1245–51. DOIPubMedGoogle Scholar
- Henderson H, Luterbach CL, Cober E, Richter SS, Salata RA, Kalayjian RC, et al. The Pitt Bacteremia Score predicts mortality in nonbacteremic infections. Clin Infect Dis. 2020;70:1826–33. DOIPubMedGoogle Scholar
- Ernst CM, Braxton JR, Rodriguez-Osorio CA, Zagieboylo AP, Li L, Pironti A, et al. Adaptive evolution of virulence and persistence in carbapenem-resistant Klebsiella pneumoniae. Nat Med. 2020;26:705–11. DOIPubMedGoogle Scholar
- Palacios M, Miner TA, Frederick DR, Sepulveda VE, Quinn JD, Walker KA, et al. Identification of two regulators of virulence that are conserved in Klebsiella pneumoniae classical and hypervirulent strains. MBio. 2018;9:e01443–18. DOIPubMedGoogle Scholar
- Prjibelski A, Antipov D, Meleshko D, Lapidus A, Korobeynikov A. Using SPAdes de novo assembler. Curr Protoc Bioinformatics. 2020;70:
e102 . DOIPubMedGoogle Scholar - Lam MMC, Wick RR, Watts SC, Cerdeira LT, Wyres KL, Holt KE. A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat Commun. 2021;12:4188. DOIPubMedGoogle Scholar
- Feldgarden M, Brover V, Gonzalez-Escalona N, Frye JG, Haendiges J, Haft DH, et al. AMRFinderPlus and the Reference Gene Catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence. Sci Rep. 2021;11:12728. DOIPubMedGoogle Scholar
- Hunt M, Mather AE, Sánchez-Busó L, Page AJ, Parkhill J, Keane JA, et al. ARIBA: rapid antimicrobial resistance genotyping directly from sequencing reads. Microb Genom. 2017;3:
e000131 . DOIPubMedGoogle Scholar - Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLOS Comput Biol. 2017;13:
e1005595 . DOIPubMedGoogle Scholar - Sullivan MJ, Petty NK, Beatson SA. Easyfig: a genome comparison visualizer. Bioinformatics. 2011;27:1009–10. DOIPubMedGoogle Scholar
- DeLeo FR, Porter AR, Kobayashi SD, Freedman B, Hao M, Jiang J, et al. Interaction of multidrug-resistant hypervirulent Klebsiella pneumoniae with components of human innate host defense. MBio. 2023;14:
e0194923 . DOIPubMedGoogle Scholar - Kobayashi SD, Voyich JM, Buhl CL, Stahl RM, DeLeo FR. Global changes in gene expression by human polymorphonuclear leukocytes during receptor-mediated phagocytosis: cell fate is regulated at the level of gene expression. Proc Natl Acad Sci U S A. 2002;99:6901–6. DOIPubMedGoogle Scholar
- Lawlor MS, Hsu J, Rick PD, Miller VL. Identification of Klebsiella pneumoniae virulence determinants using an intranasal infection model. Mol Microbiol. 2005;58:1054–73. DOIPubMedGoogle Scholar
- Tan C, Xu Z, Zheng H, Liu W, Tang X, Shou J, et al. Genome sequence of a porcine extraintestinal pathogenic Escherichia coli strain. J Bacteriol. 2011;193:5038. DOIPubMedGoogle Scholar
- Catalán-Nájera JC, Garza-Ramos U, Barrios-Camacho H. Hypervirulence and hypermucoviscosity: Two different but complementary Klebsiella spp. phenotypes? Virulence. 2017;8:1111–23. DOIPubMedGoogle Scholar
- Kochan TJ, Nozick SH, Valdes A, Mitra SD, Cheung BH, Lebrun-Corbin M, et al. Klebsiella pneumoniae clinical isolates with features of both multidrug-resistance and hypervirulence have unexpectedly low virulence. Nat Commun. 2023;14:7962. DOIPubMedGoogle Scholar
- Chen L, Chavda KD, Melano RG, Jacobs MR, Koll B, Hong T, et al. Comparative genomic analysis of KPC-encoding pKpQIL-like plasmids and their distribution in New Jersey and New York Hospitals. Antimicrob Agents Chemother. 2014;58:2871–7. DOIPubMedGoogle Scholar
- Tang N, Li Y, Yao S, Hu J, Zhao Y, Fu S, et al. Epidemicity and clonal replacement of hypervirulent carbapenem-resistant Klebsiella pneumoniae with diverse pathotypes and resistance profiles in a hospital. J Glob Antimicrob Resist. 2023;32:4–10. DOIPubMedGoogle Scholar
- Solgi H, Badmasti F, Giske CG, Aghamohammad S, Shahcheraghi F. Molecular epidemiology of NDM-1- and OXA-48-producing Klebsiella pneumoniae in an Iranian hospital: clonal dissemination of ST11 and ST893. J Antimicrob Chemother. 2018;73:1517–24. DOIPubMedGoogle Scholar
- Campbell RE, Mosimann SC, van De Rijn I, Tanner ME, Strynadka NC. The first structure of UDP-glucose dehydrogenase reveals the catalytic residues necessary for the two-fold oxidation. Biochemistry. 2000;39:7012–23. DOIPubMedGoogle Scholar
Figures
Tables
Suggested citation for this article: Jiang J, Long T, Porter AR, Lovey A, Lee A, Jacob JT, et al. Carbapenem-resistant, virulence plasmid–harboring Klebsiella pneumoniae, United States. Emerg Infect Dis. 2025 Apr [date cited]. https://doi.org/10.3201/eid3104.241396
Original Publication Date: March 11, 2025
Table of Contents – Volume 31, Number 4—April 2025
| EID Search Options |
|---|
|
|
|
|
|
|




Please use the form below to submit correspondence to the authors or contact them at the following address:
Barry N Kreiswirth, Center for Discovery and Innovation, Hackensack Meridian Health, Nutley, NJ 07110, USA
Top